
Carnobacterium inhibens subsp. gilichinskyi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae; Carnobacterium; Carnobacterium inhibens
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
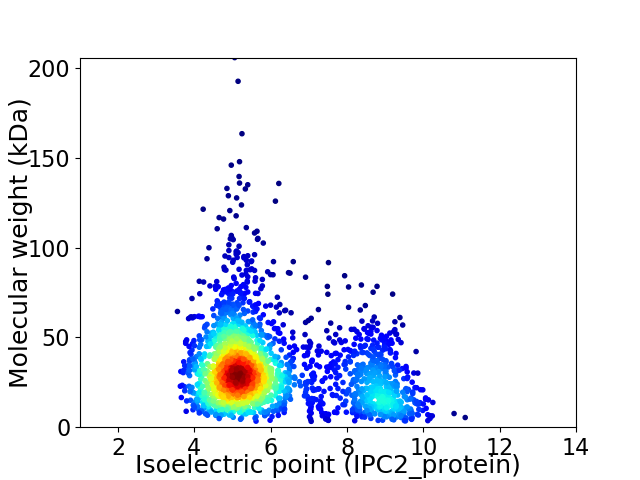
Virtual 2D-PAGE plot for 2269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5S9D6|U5S9D6_9LACT Tyrosine-protein kinase CpsD OS=Carnobacterium inhibens subsp. gilichinskyi OX=1266845 GN=Q783_06630 PE=3 SV=1
MM1 pKa = 7.54EE2 pKa = 4.96KK3 pKa = 10.53KK4 pKa = 8.46EE5 pKa = 4.1WKK7 pKa = 9.88RR8 pKa = 11.84GFIGLVSMSALLLAACGNGEE28 pKa = 4.12EE29 pKa = 4.49SNEE32 pKa = 4.18TGSTSDD38 pKa = 4.18DD39 pKa = 3.96GEE41 pKa = 4.68STTLTIDD48 pKa = 2.95VDD50 pKa = 3.84PRR52 pKa = 11.84YY53 pKa = 10.25SDD55 pKa = 3.68YY56 pKa = 11.59VNEE59 pKa = 4.86IIPAFEE65 pKa = 4.18EE66 pKa = 3.91EE67 pKa = 4.51HH68 pKa = 5.76NVKK71 pKa = 10.16IEE73 pKa = 3.5ISEE76 pKa = 4.02RR77 pKa = 11.84DD78 pKa = 3.48MFDD81 pKa = 4.2GMDD84 pKa = 4.55ALPLDD89 pKa = 4.52GPAEE93 pKa = 4.23IGSDD97 pKa = 3.34ILIAPYY103 pKa = 10.71DD104 pKa = 3.73RR105 pKa = 11.84VGTLSQQGHH114 pKa = 5.67LAEE117 pKa = 4.37VTLPDD122 pKa = 3.86DD123 pKa = 3.54EE124 pKa = 6.78RR125 pKa = 11.84YY126 pKa = 11.0DD127 pKa = 4.39EE128 pKa = 4.17MDD130 pKa = 3.12QRR132 pKa = 11.84QVTLDD137 pKa = 3.41GKK139 pKa = 10.86VYY141 pKa = 10.06GAPFVIEE148 pKa = 4.29SLVMYY153 pKa = 9.1YY154 pKa = 10.75NKK156 pKa = 10.52DD157 pKa = 3.69LIEE160 pKa = 4.06QAPEE164 pKa = 3.93TFDD167 pKa = 4.53DD168 pKa = 5.41LEE170 pKa = 4.9ALSEE174 pKa = 4.5DD175 pKa = 3.37DD176 pKa = 4.04TYY178 pKa = 11.99AFEE181 pKa = 4.63NEE183 pKa = 4.13EE184 pKa = 4.17GKK186 pKa = 8.37NTAFLANWTTAYY198 pKa = 10.5QYY200 pKa = 10.79IGLLSGYY207 pKa = 9.22GGYY210 pKa = 10.53VFGEE214 pKa = 4.33NGTDD218 pKa = 3.52PSDD221 pKa = 3.24IGLNSPEE228 pKa = 4.15SVEE231 pKa = 4.32GITYY235 pKa = 8.32ITDD238 pKa = 3.47WYY240 pKa = 9.61QNVWPQGMLDD250 pKa = 3.5ATSSEE255 pKa = 3.83NFMNEE260 pKa = 3.61LFTSGKK266 pKa = 7.09TAAVINGPWGASGYY280 pKa = 10.36EE281 pKa = 3.83EE282 pKa = 5.47AGINYY287 pKa = 8.58GVSTIPTLPSGEE299 pKa = 4.27EE300 pKa = 3.95YY301 pKa = 10.81EE302 pKa = 4.28PFAGGVAWVISKK314 pKa = 10.18YY315 pKa = 10.88SKK317 pKa = 10.84NPEE320 pKa = 4.13LAQEE324 pKa = 3.95WLDD327 pKa = 3.53YY328 pKa = 9.11VTNAEE333 pKa = 4.08NSEE336 pKa = 4.17TLYY339 pKa = 10.86EE340 pKa = 4.16LTSEE344 pKa = 4.05IPANQEE350 pKa = 2.97ARR352 pKa = 11.84TTVSEE357 pKa = 4.28TGSEE361 pKa = 3.92LSKK364 pKa = 11.21AVIEE368 pKa = 4.41QFDD371 pKa = 4.08SAVPMPNIPEE381 pKa = 4.07MTEE384 pKa = 3.48VWTGAEE390 pKa = 3.97TMIFDD395 pKa = 4.37AASGNKK401 pKa = 9.49SPQQAADD408 pKa = 3.72DD409 pKa = 3.66AVQFIEE415 pKa = 4.32EE416 pKa = 4.38NIEE419 pKa = 3.75QKK421 pKa = 10.26YY422 pKa = 8.43QNN424 pKa = 3.73
MM1 pKa = 7.54EE2 pKa = 4.96KK3 pKa = 10.53KK4 pKa = 8.46EE5 pKa = 4.1WKK7 pKa = 9.88RR8 pKa = 11.84GFIGLVSMSALLLAACGNGEE28 pKa = 4.12EE29 pKa = 4.49SNEE32 pKa = 4.18TGSTSDD38 pKa = 4.18DD39 pKa = 3.96GEE41 pKa = 4.68STTLTIDD48 pKa = 2.95VDD50 pKa = 3.84PRR52 pKa = 11.84YY53 pKa = 10.25SDD55 pKa = 3.68YY56 pKa = 11.59VNEE59 pKa = 4.86IIPAFEE65 pKa = 4.18EE66 pKa = 3.91EE67 pKa = 4.51HH68 pKa = 5.76NVKK71 pKa = 10.16IEE73 pKa = 3.5ISEE76 pKa = 4.02RR77 pKa = 11.84DD78 pKa = 3.48MFDD81 pKa = 4.2GMDD84 pKa = 4.55ALPLDD89 pKa = 4.52GPAEE93 pKa = 4.23IGSDD97 pKa = 3.34ILIAPYY103 pKa = 10.71DD104 pKa = 3.73RR105 pKa = 11.84VGTLSQQGHH114 pKa = 5.67LAEE117 pKa = 4.37VTLPDD122 pKa = 3.86DD123 pKa = 3.54EE124 pKa = 6.78RR125 pKa = 11.84YY126 pKa = 11.0DD127 pKa = 4.39EE128 pKa = 4.17MDD130 pKa = 3.12QRR132 pKa = 11.84QVTLDD137 pKa = 3.41GKK139 pKa = 10.86VYY141 pKa = 10.06GAPFVIEE148 pKa = 4.29SLVMYY153 pKa = 9.1YY154 pKa = 10.75NKK156 pKa = 10.52DD157 pKa = 3.69LIEE160 pKa = 4.06QAPEE164 pKa = 3.93TFDD167 pKa = 4.53DD168 pKa = 5.41LEE170 pKa = 4.9ALSEE174 pKa = 4.5DD175 pKa = 3.37DD176 pKa = 4.04TYY178 pKa = 11.99AFEE181 pKa = 4.63NEE183 pKa = 4.13EE184 pKa = 4.17GKK186 pKa = 8.37NTAFLANWTTAYY198 pKa = 10.5QYY200 pKa = 10.79IGLLSGYY207 pKa = 9.22GGYY210 pKa = 10.53VFGEE214 pKa = 4.33NGTDD218 pKa = 3.52PSDD221 pKa = 3.24IGLNSPEE228 pKa = 4.15SVEE231 pKa = 4.32GITYY235 pKa = 8.32ITDD238 pKa = 3.47WYY240 pKa = 9.61QNVWPQGMLDD250 pKa = 3.5ATSSEE255 pKa = 3.83NFMNEE260 pKa = 3.61LFTSGKK266 pKa = 7.09TAAVINGPWGASGYY280 pKa = 10.36EE281 pKa = 3.83EE282 pKa = 5.47AGINYY287 pKa = 8.58GVSTIPTLPSGEE299 pKa = 4.27EE300 pKa = 3.95YY301 pKa = 10.81EE302 pKa = 4.28PFAGGVAWVISKK314 pKa = 10.18YY315 pKa = 10.88SKK317 pKa = 10.84NPEE320 pKa = 4.13LAQEE324 pKa = 3.95WLDD327 pKa = 3.53YY328 pKa = 9.11VTNAEE333 pKa = 4.08NSEE336 pKa = 4.17TLYY339 pKa = 10.86EE340 pKa = 4.16LTSEE344 pKa = 4.05IPANQEE350 pKa = 2.97ARR352 pKa = 11.84TTVSEE357 pKa = 4.28TGSEE361 pKa = 3.92LSKK364 pKa = 11.21AVIEE368 pKa = 4.41QFDD371 pKa = 4.08SAVPMPNIPEE381 pKa = 4.07MTEE384 pKa = 3.48VWTGAEE390 pKa = 3.97TMIFDD395 pKa = 4.37AASGNKK401 pKa = 9.49SPQQAADD408 pKa = 3.72DD409 pKa = 3.66AVQFIEE415 pKa = 4.32EE416 pKa = 4.38NIEE419 pKa = 3.75QKK421 pKa = 10.26YY422 pKa = 8.43QNN424 pKa = 3.73
Molecular weight: 46.71 kDa
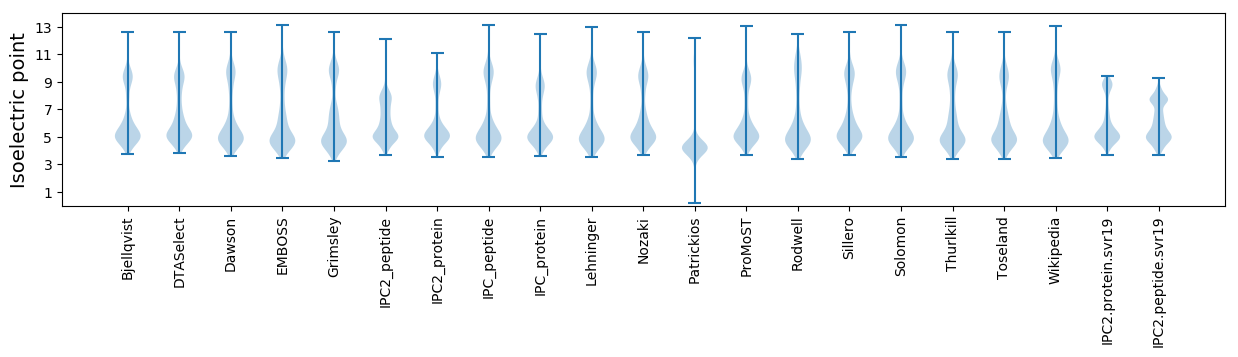
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5SBZ6|U5SBZ6_9LACT 4-hydroxy-tetrahydrodipicolinate reductase OS=Carnobacterium inhibens subsp. gilichinskyi OX=1266845 GN=dapB PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.7QPKK8 pKa = 8.83KK9 pKa = 9.05RR10 pKa = 11.84KK11 pKa = 7.43RR12 pKa = 11.84QKK14 pKa = 8.87VHH16 pKa = 5.8GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 9.83NGRR28 pKa = 11.84NVLQSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.58GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.7QPKK8 pKa = 8.83KK9 pKa = 9.05RR10 pKa = 11.84KK11 pKa = 7.43RR12 pKa = 11.84QKK14 pKa = 8.87VHH16 pKa = 5.8GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 9.83NGRR28 pKa = 11.84NVLQSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.58GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.05
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
680895 |
29 |
1792 |
300.1 |
33.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.726 ± 0.065 | 0.533 ± 0.012 |
5.417 ± 0.04 | 7.85 ± 0.053 |
4.419 ± 0.039 | 6.476 ± 0.058 |
1.784 ± 0.024 | 8.07 ± 0.053 |
7.136 ± 0.052 | 9.696 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.707 ± 0.022 | 4.842 ± 0.04 |
3.28 ± 0.025 | 3.808 ± 0.031 |
3.544 ± 0.037 | 6.346 ± 0.046 |
6.017 ± 0.043 | 6.87 ± 0.038 |
0.846 ± 0.017 | 3.634 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
