
Hubei unio douglasiae virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.78
Get precalculated fractions of proteins
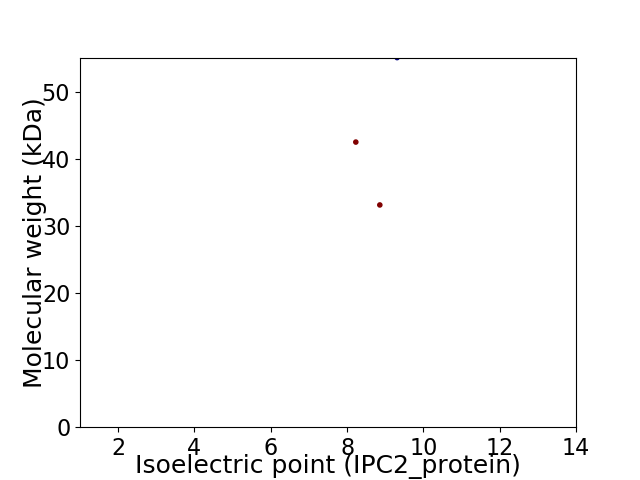
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGQ4|A0A1L3KGQ4_9VIRU RNA-directed RNA polymerase OS=Hubei unio douglasiae virus 2 OX=1923322 PE=4 SV=1
MM1 pKa = 7.27FLKK4 pKa = 10.37DD5 pKa = 4.07DD6 pKa = 5.19KK7 pKa = 9.77YY8 pKa = 11.17HH9 pKa = 5.53EE10 pKa = 4.53WKK12 pKa = 10.08PVAPRR17 pKa = 11.84CIQYY21 pKa = 10.03RR22 pKa = 11.84GKK24 pKa = 10.69RR25 pKa = 11.84YY26 pKa = 9.77ALSLACYY33 pKa = 8.36LHH35 pKa = 7.2PIEE38 pKa = 4.37QMVYY42 pKa = 9.32GWTDD46 pKa = 3.29LSGTPIFAKK55 pKa = 10.31SRR57 pKa = 11.84NLTQRR62 pKa = 11.84GHH64 pKa = 7.42DD65 pKa = 3.38IADD68 pKa = 3.41KK69 pKa = 10.38MSYY72 pKa = 9.16FQNPVAISLDD82 pKa = 3.37HH83 pKa = 6.84SKK85 pKa = 10.9FDD87 pKa = 3.31AHH89 pKa = 8.2VNMSLLDD96 pKa = 3.86LEE98 pKa = 4.2HH99 pKa = 7.04WFYY102 pKa = 11.12KK103 pKa = 9.3QCNRR107 pKa = 11.84SPKK110 pKa = 10.54LKK112 pKa = 9.88MLLHH116 pKa = 6.07WQRR119 pKa = 11.84VNHH122 pKa = 5.86GQTKK126 pKa = 10.49NGTKK130 pKa = 10.11YY131 pKa = 7.79VTRR134 pKa = 11.84ATRR137 pKa = 11.84MSGDD141 pKa = 3.32QNTGIGNSIINYY153 pKa = 10.1AMTKK157 pKa = 10.66ALLEE161 pKa = 4.01KK162 pKa = 10.73LKK164 pKa = 10.33IRR166 pKa = 11.84HH167 pKa = 6.24CLYY170 pKa = 10.31IDD172 pKa = 3.92GDD174 pKa = 4.27DD175 pKa = 3.94FLVICEE181 pKa = 4.87RR182 pKa = 11.84IDD184 pKa = 3.3AGKK187 pKa = 10.22FDD189 pKa = 4.36PSLYY193 pKa = 9.96KK194 pKa = 10.38QFGMATKK201 pKa = 9.9LDD203 pKa = 4.46SITGVIEE210 pKa = 5.59HH211 pKa = 7.43IDD213 pKa = 3.6FCQCRR218 pKa = 11.84PVFNGSGYY226 pKa = 9.43TMVRR230 pKa = 11.84DD231 pKa = 4.91PYY233 pKa = 11.52RR234 pKa = 11.84MLQRR238 pKa = 11.84IQWAVGKK245 pKa = 9.37KK246 pKa = 9.71HH247 pKa = 6.11PRR249 pKa = 11.84HH250 pKa = 4.89VVNYY254 pKa = 6.82LTSIGKK260 pKa = 9.76CMIALGMGLPVEE272 pKa = 4.27QYY274 pKa = 10.39IGSTLSNLGGKK285 pKa = 10.05YY286 pKa = 8.35ITTEE290 pKa = 3.67QTMMANKK297 pKa = 9.54MFMRR301 pKa = 11.84PSNARR306 pKa = 11.84VVEE309 pKa = 4.18CCEE312 pKa = 3.85ASRR315 pKa = 11.84ASYY318 pKa = 8.47EE319 pKa = 4.17QAWGFTVGQQLILEE333 pKa = 4.58RR334 pKa = 11.84LSIGPPVLEE343 pKa = 4.77DD344 pKa = 3.69LVPFLQYY351 pKa = 10.92GSEE354 pKa = 4.08EE355 pKa = 4.46TVTEE359 pKa = 4.73GIRR362 pKa = 11.84PFSSSPIGSVYY373 pKa = 11.04
MM1 pKa = 7.27FLKK4 pKa = 10.37DD5 pKa = 4.07DD6 pKa = 5.19KK7 pKa = 9.77YY8 pKa = 11.17HH9 pKa = 5.53EE10 pKa = 4.53WKK12 pKa = 10.08PVAPRR17 pKa = 11.84CIQYY21 pKa = 10.03RR22 pKa = 11.84GKK24 pKa = 10.69RR25 pKa = 11.84YY26 pKa = 9.77ALSLACYY33 pKa = 8.36LHH35 pKa = 7.2PIEE38 pKa = 4.37QMVYY42 pKa = 9.32GWTDD46 pKa = 3.29LSGTPIFAKK55 pKa = 10.31SRR57 pKa = 11.84NLTQRR62 pKa = 11.84GHH64 pKa = 7.42DD65 pKa = 3.38IADD68 pKa = 3.41KK69 pKa = 10.38MSYY72 pKa = 9.16FQNPVAISLDD82 pKa = 3.37HH83 pKa = 6.84SKK85 pKa = 10.9FDD87 pKa = 3.31AHH89 pKa = 8.2VNMSLLDD96 pKa = 3.86LEE98 pKa = 4.2HH99 pKa = 7.04WFYY102 pKa = 11.12KK103 pKa = 9.3QCNRR107 pKa = 11.84SPKK110 pKa = 10.54LKK112 pKa = 9.88MLLHH116 pKa = 6.07WQRR119 pKa = 11.84VNHH122 pKa = 5.86GQTKK126 pKa = 10.49NGTKK130 pKa = 10.11YY131 pKa = 7.79VTRR134 pKa = 11.84ATRR137 pKa = 11.84MSGDD141 pKa = 3.32QNTGIGNSIINYY153 pKa = 10.1AMTKK157 pKa = 10.66ALLEE161 pKa = 4.01KK162 pKa = 10.73LKK164 pKa = 10.33IRR166 pKa = 11.84HH167 pKa = 6.24CLYY170 pKa = 10.31IDD172 pKa = 3.92GDD174 pKa = 4.27DD175 pKa = 3.94FLVICEE181 pKa = 4.87RR182 pKa = 11.84IDD184 pKa = 3.3AGKK187 pKa = 10.22FDD189 pKa = 4.36PSLYY193 pKa = 9.96KK194 pKa = 10.38QFGMATKK201 pKa = 9.9LDD203 pKa = 4.46SITGVIEE210 pKa = 5.59HH211 pKa = 7.43IDD213 pKa = 3.6FCQCRR218 pKa = 11.84PVFNGSGYY226 pKa = 9.43TMVRR230 pKa = 11.84DD231 pKa = 4.91PYY233 pKa = 11.52RR234 pKa = 11.84MLQRR238 pKa = 11.84IQWAVGKK245 pKa = 9.37KK246 pKa = 9.71HH247 pKa = 6.11PRR249 pKa = 11.84HH250 pKa = 4.89VVNYY254 pKa = 6.82LTSIGKK260 pKa = 9.76CMIALGMGLPVEE272 pKa = 4.27QYY274 pKa = 10.39IGSTLSNLGGKK285 pKa = 10.05YY286 pKa = 8.35ITTEE290 pKa = 3.67QTMMANKK297 pKa = 9.54MFMRR301 pKa = 11.84PSNARR306 pKa = 11.84VVEE309 pKa = 4.18CCEE312 pKa = 3.85ASRR315 pKa = 11.84ASYY318 pKa = 8.47EE319 pKa = 4.17QAWGFTVGQQLILEE333 pKa = 4.58RR334 pKa = 11.84LSIGPPVLEE343 pKa = 4.77DD344 pKa = 3.69LVPFLQYY351 pKa = 10.92GSEE354 pKa = 4.08EE355 pKa = 4.46TVTEE359 pKa = 4.73GIRR362 pKa = 11.84PFSSSPIGSVYY373 pKa = 11.04
Molecular weight: 42.56 kDa
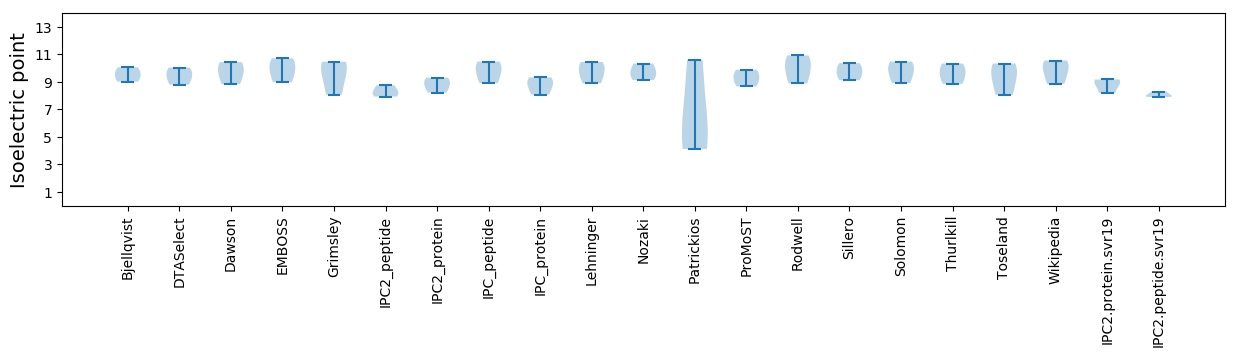
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGQ4|A0A1L3KGQ4_9VIRU RNA-directed RNA polymerase OS=Hubei unio douglasiae virus 2 OX=1923322 PE=4 SV=1
MM1 pKa = 7.66SSPTVKK7 pKa = 10.42SVVPNAGGNASVTGTGGLDD26 pKa = 3.12HH27 pKa = 6.99PTPGRR32 pKa = 11.84TGIRR36 pKa = 11.84ARR38 pKa = 11.84SGEE41 pKa = 4.16PGHH44 pKa = 6.49GRR46 pKa = 11.84NVCNRR51 pKa = 11.84KK52 pKa = 6.95TASRR56 pKa = 11.84RR57 pKa = 11.84NNEE60 pKa = 3.8EE61 pKa = 3.98PIKK64 pKa = 10.68RR65 pKa = 11.84SAPPNRR71 pKa = 11.84RR72 pKa = 11.84EE73 pKa = 4.19GRR75 pKa = 11.84DD76 pKa = 3.33KK77 pKa = 11.12RR78 pKa = 11.84PAGVPLGKK86 pKa = 10.6GKK88 pKa = 10.42GRR90 pKa = 11.84EE91 pKa = 4.22CGTLAPYY98 pKa = 10.38ARR100 pKa = 11.84SQQGRR105 pKa = 11.84APRR108 pKa = 11.84EE109 pKa = 3.75KK110 pKa = 10.49ASNQGFHH117 pKa = 6.94PKK119 pKa = 9.72EE120 pKa = 4.18DD121 pKa = 5.72PIPPWNRR128 pKa = 11.84VVNRR132 pKa = 11.84RR133 pKa = 11.84SDD135 pKa = 3.41RR136 pKa = 11.84AKK138 pKa = 10.97LLILIQQLNLRR149 pKa = 11.84IIQLEE154 pKa = 4.21RR155 pKa = 11.84EE156 pKa = 4.42AMRR159 pKa = 11.84LPSGQFQSAPILLARR174 pKa = 11.84NGWSTPASSFQASPKK189 pKa = 10.14LSRR192 pKa = 11.84NDD194 pKa = 2.84KK195 pKa = 10.07SEE197 pKa = 4.02RR198 pKa = 11.84KK199 pKa = 9.65RR200 pKa = 11.84EE201 pKa = 3.98VRR203 pKa = 11.84DD204 pKa = 3.64SNCEE208 pKa = 3.69VKK210 pKa = 10.87QNVLPPQPDD219 pKa = 3.23AQASGEE225 pKa = 4.3STAPQPTKK233 pKa = 10.06TPSPAGSRR241 pKa = 11.84KK242 pKa = 9.43SNRR245 pKa = 11.84GDD247 pKa = 3.15PTTPSSRR254 pKa = 11.84RR255 pKa = 11.84SRR257 pKa = 11.84KK258 pKa = 8.63SSYY261 pKa = 10.69GGSCEE266 pKa = 3.74TSAPFVGAKK275 pKa = 8.41TATEE279 pKa = 4.27PPSDD283 pKa = 3.34HH284 pKa = 6.9SSYY287 pKa = 10.94HH288 pKa = 6.57RR289 pKa = 11.84DD290 pKa = 3.16ISCTSAASTVSGVSRR305 pKa = 11.84CMSDD309 pKa = 2.9LTSEE313 pKa = 4.56EE314 pKa = 4.23KK315 pKa = 10.38PSSKK319 pKa = 9.86EE320 pKa = 3.79HH321 pKa = 6.04SRR323 pKa = 11.84KK324 pKa = 9.1PPKK327 pKa = 10.05NAGRR331 pKa = 11.84SYY333 pKa = 10.8QWTKK337 pKa = 10.84EE338 pKa = 4.1SGPMDD343 pKa = 4.68ANLDD347 pKa = 4.12PIYY350 pKa = 10.41GAEE353 pKa = 4.07VKK355 pKa = 10.03EE356 pKa = 4.23IPRR359 pKa = 11.84GRR361 pKa = 11.84VKK363 pKa = 10.74KK364 pKa = 10.57SIRR367 pKa = 11.84KK368 pKa = 7.36WNKK371 pKa = 4.52TTKK374 pKa = 10.1VLPTEE379 pKa = 4.43VEE381 pKa = 3.88LHH383 pKa = 6.49FYY385 pKa = 10.83LVLEE389 pKa = 4.5FAFVPRR395 pKa = 11.84DD396 pKa = 3.83CTVMRR401 pKa = 11.84QMTNKK406 pKa = 9.92AKK408 pKa = 10.57SYY410 pKa = 11.05LNTFDD415 pKa = 3.36TSNYY419 pKa = 6.94TAEE422 pKa = 3.86EE423 pKa = 4.12RR424 pKa = 11.84FRR426 pKa = 11.84LVMKK430 pKa = 10.35AVRR433 pKa = 11.84SAMCVTEE440 pKa = 4.35EE441 pKa = 4.3EE442 pKa = 4.44EE443 pKa = 4.56CVRR446 pKa = 11.84AALKK450 pKa = 11.05NPDD453 pKa = 3.71VQEE456 pKa = 4.38LAHH459 pKa = 6.4KK460 pKa = 9.83QAAFIQEE467 pKa = 4.02GFVGNEE473 pKa = 4.22GIGSVKK479 pKa = 10.45SYY481 pKa = 11.03LSNACKK487 pKa = 10.33INVFKK492 pKa = 11.03KK493 pKa = 9.47GTHH496 pKa = 5.59LPKK499 pKa = 10.17KK500 pKa = 10.22VKK502 pKa = 10.37
MM1 pKa = 7.66SSPTVKK7 pKa = 10.42SVVPNAGGNASVTGTGGLDD26 pKa = 3.12HH27 pKa = 6.99PTPGRR32 pKa = 11.84TGIRR36 pKa = 11.84ARR38 pKa = 11.84SGEE41 pKa = 4.16PGHH44 pKa = 6.49GRR46 pKa = 11.84NVCNRR51 pKa = 11.84KK52 pKa = 6.95TASRR56 pKa = 11.84RR57 pKa = 11.84NNEE60 pKa = 3.8EE61 pKa = 3.98PIKK64 pKa = 10.68RR65 pKa = 11.84SAPPNRR71 pKa = 11.84RR72 pKa = 11.84EE73 pKa = 4.19GRR75 pKa = 11.84DD76 pKa = 3.33KK77 pKa = 11.12RR78 pKa = 11.84PAGVPLGKK86 pKa = 10.6GKK88 pKa = 10.42GRR90 pKa = 11.84EE91 pKa = 4.22CGTLAPYY98 pKa = 10.38ARR100 pKa = 11.84SQQGRR105 pKa = 11.84APRR108 pKa = 11.84EE109 pKa = 3.75KK110 pKa = 10.49ASNQGFHH117 pKa = 6.94PKK119 pKa = 9.72EE120 pKa = 4.18DD121 pKa = 5.72PIPPWNRR128 pKa = 11.84VVNRR132 pKa = 11.84RR133 pKa = 11.84SDD135 pKa = 3.41RR136 pKa = 11.84AKK138 pKa = 10.97LLILIQQLNLRR149 pKa = 11.84IIQLEE154 pKa = 4.21RR155 pKa = 11.84EE156 pKa = 4.42AMRR159 pKa = 11.84LPSGQFQSAPILLARR174 pKa = 11.84NGWSTPASSFQASPKK189 pKa = 10.14LSRR192 pKa = 11.84NDD194 pKa = 2.84KK195 pKa = 10.07SEE197 pKa = 4.02RR198 pKa = 11.84KK199 pKa = 9.65RR200 pKa = 11.84EE201 pKa = 3.98VRR203 pKa = 11.84DD204 pKa = 3.64SNCEE208 pKa = 3.69VKK210 pKa = 10.87QNVLPPQPDD219 pKa = 3.23AQASGEE225 pKa = 4.3STAPQPTKK233 pKa = 10.06TPSPAGSRR241 pKa = 11.84KK242 pKa = 9.43SNRR245 pKa = 11.84GDD247 pKa = 3.15PTTPSSRR254 pKa = 11.84RR255 pKa = 11.84SRR257 pKa = 11.84KK258 pKa = 8.63SSYY261 pKa = 10.69GGSCEE266 pKa = 3.74TSAPFVGAKK275 pKa = 8.41TATEE279 pKa = 4.27PPSDD283 pKa = 3.34HH284 pKa = 6.9SSYY287 pKa = 10.94HH288 pKa = 6.57RR289 pKa = 11.84DD290 pKa = 3.16ISCTSAASTVSGVSRR305 pKa = 11.84CMSDD309 pKa = 2.9LTSEE313 pKa = 4.56EE314 pKa = 4.23KK315 pKa = 10.38PSSKK319 pKa = 9.86EE320 pKa = 3.79HH321 pKa = 6.04SRR323 pKa = 11.84KK324 pKa = 9.1PPKK327 pKa = 10.05NAGRR331 pKa = 11.84SYY333 pKa = 10.8QWTKK337 pKa = 10.84EE338 pKa = 4.1SGPMDD343 pKa = 4.68ANLDD347 pKa = 4.12PIYY350 pKa = 10.41GAEE353 pKa = 4.07VKK355 pKa = 10.03EE356 pKa = 4.23IPRR359 pKa = 11.84GRR361 pKa = 11.84VKK363 pKa = 10.74KK364 pKa = 10.57SIRR367 pKa = 11.84KK368 pKa = 7.36WNKK371 pKa = 4.52TTKK374 pKa = 10.1VLPTEE379 pKa = 4.43VEE381 pKa = 3.88LHH383 pKa = 6.49FYY385 pKa = 10.83LVLEE389 pKa = 4.5FAFVPRR395 pKa = 11.84DD396 pKa = 3.83CTVMRR401 pKa = 11.84QMTNKK406 pKa = 9.92AKK408 pKa = 10.57SYY410 pKa = 11.05LNTFDD415 pKa = 3.36TSNYY419 pKa = 6.94TAEE422 pKa = 3.86EE423 pKa = 4.12RR424 pKa = 11.84FRR426 pKa = 11.84LVMKK430 pKa = 10.35AVRR433 pKa = 11.84SAMCVTEE440 pKa = 4.35EE441 pKa = 4.3EE442 pKa = 4.44EE443 pKa = 4.56CVRR446 pKa = 11.84AALKK450 pKa = 11.05NPDD453 pKa = 3.71VQEE456 pKa = 4.38LAHH459 pKa = 6.4KK460 pKa = 9.83QAAFIQEE467 pKa = 4.02GFVGNEE473 pKa = 4.22GIGSVKK479 pKa = 10.45SYY481 pKa = 11.03LSNACKK487 pKa = 10.33INVFKK492 pKa = 11.03KK493 pKa = 9.47GTHH496 pKa = 5.59LPKK499 pKa = 10.17KK500 pKa = 10.22VKK502 pKa = 10.37
Molecular weight: 55.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1182 |
307 |
502 |
394.0 |
43.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.276 ± 0.898 | 1.946 ± 0.371 |
3.807 ± 0.417 | 5.076 ± 0.823 |
3.299 ± 0.502 | 7.783 ± 0.49 |
1.946 ± 0.557 | 4.315 ± 0.875 |
6.345 ± 1.016 | 6.514 ± 0.861 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.876 ± 0.711 | 4.653 ± 0.364 |
6.853 ± 0.964 | 4.315 ± 0.391 |
7.191 ± 1.054 | 8.883 ± 0.984 |
6.43 ± 0.725 | 6.176 ± 0.354 |
1.438 ± 0.379 | 2.876 ± 0.871 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
