
Canine coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Tegacovirus; Alphacoronavirus 1
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
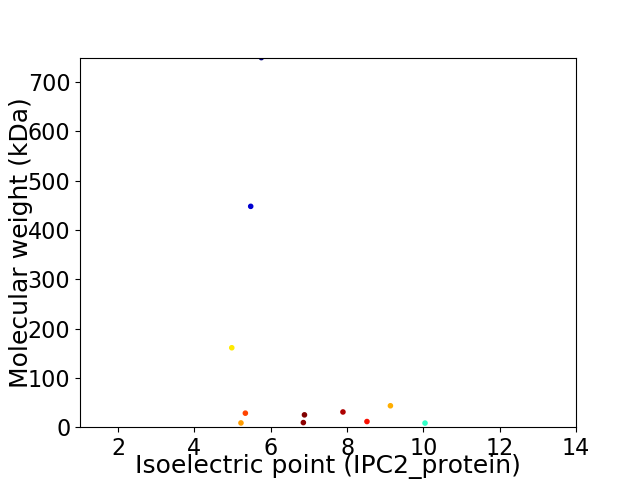
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

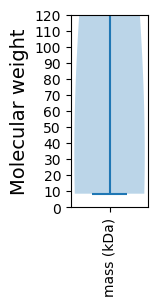
Protein with the lowest isoelectric point:
>tr|H9TEX6|H9TEX6_9ALPC Membrane protein OS=Canine coronavirus OX=11153 GN=M PE=3 SV=1
MM1 pKa = 7.4IVLVTCILLLCSYY14 pKa = 9.04HH15 pKa = 5.95TVLSTANNDD24 pKa = 3.13CRR26 pKa = 11.84QVNVTQLPGNEE37 pKa = 3.63YY38 pKa = 10.62LIRR41 pKa = 11.84DD42 pKa = 3.96FLFQSFKK49 pKa = 11.11EE50 pKa = 4.16EE51 pKa = 4.09GSVVVGGYY59 pKa = 10.2YY60 pKa = 8.53PTEE63 pKa = 3.31VWYY66 pKa = 10.62NCSRR70 pKa = 11.84TATTTAYY77 pKa = 10.43QYY79 pKa = 11.1FNNIHH84 pKa = 6.68AFYY87 pKa = 10.77FDD89 pKa = 3.66MEE91 pKa = 4.25AMEE94 pKa = 4.6NSTGNARR101 pKa = 11.84GKK103 pKa = 9.27PLLFHH108 pKa = 5.86VHH110 pKa = 6.68GDD112 pKa = 3.65PVSVIIYY119 pKa = 9.4ISAYY123 pKa = 9.94RR124 pKa = 11.84DD125 pKa = 3.89DD126 pKa = 4.15VQHH129 pKa = 7.2RR130 pKa = 11.84PLLKK134 pKa = 10.4HH135 pKa = 6.16GLVCITKK142 pKa = 10.19SRR144 pKa = 11.84NIDD147 pKa = 3.41YY148 pKa = 7.88NTFTSSQWNSICTGNDD164 pKa = 2.5RR165 pKa = 11.84KK166 pKa = 10.45IPFSVIPTDD175 pKa = 3.44NGTKK179 pKa = 9.62IYY181 pKa = 10.47GLEE184 pKa = 4.03WNDD187 pKa = 3.62EE188 pKa = 4.1FVTAYY193 pKa = 9.89ISGHH197 pKa = 5.06SYY199 pKa = 10.18NWNINNNWFNNVTLLYY215 pKa = 10.18SRR217 pKa = 11.84SSTATWQHH225 pKa = 4.71SAAYY229 pKa = 9.59VYY231 pKa = 10.56QGVSNFTYY239 pKa = 10.84YY240 pKa = 11.24KK241 pKa = 10.71LNNTNGLKK249 pKa = 9.77TYY251 pKa = 7.25EE252 pKa = 3.93FCEE255 pKa = 4.49DD256 pKa = 3.71YY257 pKa = 10.67EE258 pKa = 4.64YY259 pKa = 10.83CTATATNVFAPTVGGYY275 pKa = 9.69IPDD278 pKa = 3.64GFSFNNWFLLTNSSTFVSGRR298 pKa = 11.84FVTNQPLLVNCLWPVPSFGVAAQEE322 pKa = 4.3FCFEE326 pKa = 4.3GAQFSQCNGMSLNNTVDD343 pKa = 3.78VIRR346 pKa = 11.84FNLNFTADD354 pKa = 3.63VQSGMGATVFSLNTTGGVILEE375 pKa = 4.07ISCYY379 pKa = 9.9KK380 pKa = 10.17DD381 pKa = 3.22TVSEE385 pKa = 4.31SSFYY389 pKa = 10.93SYY391 pKa = 11.79GEE393 pKa = 3.88IPFGITDD400 pKa = 4.33GPRR403 pKa = 11.84YY404 pKa = 10.15CYY406 pKa = 10.84VLYY409 pKa = 10.96NGTALKK415 pKa = 10.82YY416 pKa = 10.58LGTLPPSVKK425 pKa = 10.2EE426 pKa = 3.64IAISKK431 pKa = 8.76WGHH434 pKa = 5.86FYY436 pKa = 10.78INGYY440 pKa = 9.41NFFSTFPIDD449 pKa = 4.28CISFNLTTGASGAFWTIAYY468 pKa = 7.76TSYY471 pKa = 11.12TEE473 pKa = 5.09ALVQVEE479 pKa = 4.01NTAIKK484 pKa = 10.3KK485 pKa = 7.68VTYY488 pKa = 10.28CNSHH492 pKa = 6.35INNIKK497 pKa = 10.33CSQLTANLNNGFYY510 pKa = 10.08PVASSEE516 pKa = 4.07VGLVNKK522 pKa = 10.31SVVLLPTFFAHH533 pKa = 5.84TAFNITIDD541 pKa = 3.66LGMKK545 pKa = 10.04RR546 pKa = 11.84SGYY549 pKa = 8.22GQPIASTLSNITLPMQDD566 pKa = 3.7NNTDD570 pKa = 3.69VYY572 pKa = 10.5CVRR575 pKa = 11.84SNQFSVYY582 pKa = 8.11VHH584 pKa = 5.76STCKK588 pKa = 10.33SSSWDD593 pKa = 3.38NIFNQDD599 pKa = 3.41CTDD602 pKa = 3.44VLEE605 pKa = 4.39ATAVIKK611 pKa = 9.94TGTCPFSFDD620 pKa = 3.91KK621 pKa = 11.18LNNYY625 pKa = 7.76LTFNKK630 pKa = 9.63FCLSLSPVGANCKK643 pKa = 9.97FDD645 pKa = 3.74VAAHH649 pKa = 5.81TRR651 pKa = 11.84TNEE654 pKa = 3.69QVVRR658 pKa = 11.84SLYY661 pKa = 10.45VIYY664 pKa = 10.55EE665 pKa = 4.03EE666 pKa = 4.75GDD668 pKa = 3.7NIVGVPSDD676 pKa = 3.48NSGLHH681 pKa = 6.34DD682 pKa = 5.3LSVLHH687 pKa = 7.02LDD689 pKa = 3.23SCTDD693 pKa = 3.23YY694 pKa = 11.33NIYY697 pKa = 10.59GRR699 pKa = 11.84TGVGIIRR706 pKa = 11.84QTNSTLLSGLYY717 pKa = 8.14YY718 pKa = 10.47TSLSGDD724 pKa = 3.35LLGFKK729 pKa = 10.1NVSDD733 pKa = 3.82GVIYY737 pKa = 10.67SVTPCDD743 pKa = 3.31VSAQVAVIDD752 pKa = 4.2GAIVGAMTSINSEE765 pKa = 4.32LLGLTHH771 pKa = 7.13WTTTPNFYY779 pKa = 10.7YY780 pKa = 10.66YY781 pKa = 10.66SIYY784 pKa = 10.78NYY786 pKa = 9.49TNEE789 pKa = 4.22RR790 pKa = 11.84TRR792 pKa = 11.84GTAIDD797 pKa = 4.46SNDD800 pKa = 3.38VDD802 pKa = 4.54CEE804 pKa = 4.34PIITYY809 pKa = 11.09ANIGVCKK816 pKa = 10.5NGALVFINVTHH827 pKa = 7.03SDD829 pKa = 3.31GDD831 pKa = 3.76VQPISTGNVTIPTNFTISVQVEE853 pKa = 4.08YY854 pKa = 10.31IQVYY858 pKa = 5.57TTPVSIDD865 pKa = 3.21CSRR868 pKa = 11.84YY869 pKa = 8.47VCNGNPRR876 pKa = 11.84CNKK879 pKa = 10.11LLTQYY884 pKa = 10.84VSACQTIEE892 pKa = 3.6QALAMGARR900 pKa = 11.84LEE902 pKa = 4.0NMEE905 pKa = 4.38VDD907 pKa = 3.28SMLFVSEE914 pKa = 4.35NALKK918 pKa = 10.36LASVEE923 pKa = 4.2AFNSTEE929 pKa = 3.91NLDD932 pKa = 4.31PIYY935 pKa = 10.67KK936 pKa = 9.41EE937 pKa = 3.84WPNIGGSWLGGLKK950 pKa = 10.5DD951 pKa = 4.17ILPSHH956 pKa = 5.68NSKK959 pKa = 10.42RR960 pKa = 11.84KK961 pKa = 8.03YY962 pKa = 10.0RR963 pKa = 11.84SAIEE967 pKa = 4.79DD968 pKa = 3.77LLFDD972 pKa = 3.96KK973 pKa = 11.1VVTSGLGTVDD983 pKa = 3.79EE984 pKa = 4.94DD985 pKa = 4.2YY986 pKa = 11.18KK987 pKa = 11.08RR988 pKa = 11.84CTGGYY993 pKa = 10.17DD994 pKa = 3.45IADD997 pKa = 4.27LVCAQYY1003 pKa = 11.58YY1004 pKa = 8.99NGIMVLPGVANDD1016 pKa = 5.23DD1017 pKa = 3.64KK1018 pKa = 10.09MTMYY1022 pKa = 9.25TASLAGGITLGALGGGAVAIPFAVAVQARR1051 pKa = 11.84LNYY1054 pKa = 9.72VALQTDD1060 pKa = 4.12VLNKK1064 pKa = 9.83NQQILANAFNQAIGNITQAFGKK1086 pKa = 10.65VNDD1089 pKa = 5.6AIHH1092 pKa = 5.36QTSQGLATVAKK1103 pKa = 10.13ALAKK1107 pKa = 10.36VQDD1110 pKa = 3.83VVNTQGQALSHH1121 pKa = 6.03LTVQLQNNFQAISSSISDD1139 pKa = 3.33IYY1141 pKa = 11.39NRR1143 pKa = 11.84LDD1145 pKa = 3.61EE1146 pKa = 4.93LSADD1150 pKa = 3.68AQVDD1154 pKa = 3.66RR1155 pKa = 11.84LITGRR1160 pKa = 11.84LTALNAFVSQTLTRR1174 pKa = 11.84QAEE1177 pKa = 4.39VRR1179 pKa = 11.84ASRR1182 pKa = 11.84QLAKK1186 pKa = 10.86DD1187 pKa = 3.4KK1188 pKa = 11.38VNEE1191 pKa = 4.27CVRR1194 pKa = 11.84SQSQRR1199 pKa = 11.84FGFCGNGTHH1208 pKa = 7.49LFSLANAAPNGMIFFHH1224 pKa = 6.14TVLLPTAYY1232 pKa = 8.01EE1233 pKa = 4.33TVTAWSGICASDD1245 pKa = 3.56GDD1247 pKa = 4.1RR1248 pKa = 11.84TFGLVVKK1255 pKa = 10.13DD1256 pKa = 3.57VQLTLFRR1263 pKa = 11.84NLDD1266 pKa = 3.49GKK1268 pKa = 11.0FYY1270 pKa = 9.4LTPRR1274 pKa = 11.84TMYY1277 pKa = 9.68QPRR1280 pKa = 11.84VVTSSDD1286 pKa = 3.09FVQIEE1291 pKa = 4.46GCDD1294 pKa = 3.54VLFVNATVIDD1304 pKa = 4.59LPSIIPDD1311 pKa = 3.84YY1312 pKa = 11.09IDD1314 pKa = 3.75INQTVQDD1321 pKa = 3.37ILEE1324 pKa = 4.31NYY1326 pKa = 8.5RR1327 pKa = 11.84PNWTVPEE1334 pKa = 4.11LTLDD1338 pKa = 3.76IFNATYY1344 pKa = 11.19LNLTGEE1350 pKa = 4.13IDD1352 pKa = 3.65DD1353 pKa = 5.06LEE1355 pKa = 4.33FRR1357 pKa = 11.84SEE1359 pKa = 4.1KK1360 pKa = 10.53LHH1362 pKa = 5.34NTTVEE1367 pKa = 3.84LAILIDD1373 pKa = 4.43NINNTLVNLEE1383 pKa = 3.92WLNRR1387 pKa = 11.84IEE1389 pKa = 5.6TYY1391 pKa = 10.75VKK1393 pKa = 9.1WPWYY1397 pKa = 8.39VWLLIGLVVIFCIPLLLFCCCSTGCCGCIGCLGSCCHH1434 pKa = 6.85SICSRR1439 pKa = 11.84RR1440 pKa = 11.84QFEE1443 pKa = 4.2NYY1445 pKa = 9.36EE1446 pKa = 4.38PIEE1449 pKa = 4.26KK1450 pKa = 10.17VHH1452 pKa = 5.03VHH1454 pKa = 6.07
MM1 pKa = 7.4IVLVTCILLLCSYY14 pKa = 9.04HH15 pKa = 5.95TVLSTANNDD24 pKa = 3.13CRR26 pKa = 11.84QVNVTQLPGNEE37 pKa = 3.63YY38 pKa = 10.62LIRR41 pKa = 11.84DD42 pKa = 3.96FLFQSFKK49 pKa = 11.11EE50 pKa = 4.16EE51 pKa = 4.09GSVVVGGYY59 pKa = 10.2YY60 pKa = 8.53PTEE63 pKa = 3.31VWYY66 pKa = 10.62NCSRR70 pKa = 11.84TATTTAYY77 pKa = 10.43QYY79 pKa = 11.1FNNIHH84 pKa = 6.68AFYY87 pKa = 10.77FDD89 pKa = 3.66MEE91 pKa = 4.25AMEE94 pKa = 4.6NSTGNARR101 pKa = 11.84GKK103 pKa = 9.27PLLFHH108 pKa = 5.86VHH110 pKa = 6.68GDD112 pKa = 3.65PVSVIIYY119 pKa = 9.4ISAYY123 pKa = 9.94RR124 pKa = 11.84DD125 pKa = 3.89DD126 pKa = 4.15VQHH129 pKa = 7.2RR130 pKa = 11.84PLLKK134 pKa = 10.4HH135 pKa = 6.16GLVCITKK142 pKa = 10.19SRR144 pKa = 11.84NIDD147 pKa = 3.41YY148 pKa = 7.88NTFTSSQWNSICTGNDD164 pKa = 2.5RR165 pKa = 11.84KK166 pKa = 10.45IPFSVIPTDD175 pKa = 3.44NGTKK179 pKa = 9.62IYY181 pKa = 10.47GLEE184 pKa = 4.03WNDD187 pKa = 3.62EE188 pKa = 4.1FVTAYY193 pKa = 9.89ISGHH197 pKa = 5.06SYY199 pKa = 10.18NWNINNNWFNNVTLLYY215 pKa = 10.18SRR217 pKa = 11.84SSTATWQHH225 pKa = 4.71SAAYY229 pKa = 9.59VYY231 pKa = 10.56QGVSNFTYY239 pKa = 10.84YY240 pKa = 11.24KK241 pKa = 10.71LNNTNGLKK249 pKa = 9.77TYY251 pKa = 7.25EE252 pKa = 3.93FCEE255 pKa = 4.49DD256 pKa = 3.71YY257 pKa = 10.67EE258 pKa = 4.64YY259 pKa = 10.83CTATATNVFAPTVGGYY275 pKa = 9.69IPDD278 pKa = 3.64GFSFNNWFLLTNSSTFVSGRR298 pKa = 11.84FVTNQPLLVNCLWPVPSFGVAAQEE322 pKa = 4.3FCFEE326 pKa = 4.3GAQFSQCNGMSLNNTVDD343 pKa = 3.78VIRR346 pKa = 11.84FNLNFTADD354 pKa = 3.63VQSGMGATVFSLNTTGGVILEE375 pKa = 4.07ISCYY379 pKa = 9.9KK380 pKa = 10.17DD381 pKa = 3.22TVSEE385 pKa = 4.31SSFYY389 pKa = 10.93SYY391 pKa = 11.79GEE393 pKa = 3.88IPFGITDD400 pKa = 4.33GPRR403 pKa = 11.84YY404 pKa = 10.15CYY406 pKa = 10.84VLYY409 pKa = 10.96NGTALKK415 pKa = 10.82YY416 pKa = 10.58LGTLPPSVKK425 pKa = 10.2EE426 pKa = 3.64IAISKK431 pKa = 8.76WGHH434 pKa = 5.86FYY436 pKa = 10.78INGYY440 pKa = 9.41NFFSTFPIDD449 pKa = 4.28CISFNLTTGASGAFWTIAYY468 pKa = 7.76TSYY471 pKa = 11.12TEE473 pKa = 5.09ALVQVEE479 pKa = 4.01NTAIKK484 pKa = 10.3KK485 pKa = 7.68VTYY488 pKa = 10.28CNSHH492 pKa = 6.35INNIKK497 pKa = 10.33CSQLTANLNNGFYY510 pKa = 10.08PVASSEE516 pKa = 4.07VGLVNKK522 pKa = 10.31SVVLLPTFFAHH533 pKa = 5.84TAFNITIDD541 pKa = 3.66LGMKK545 pKa = 10.04RR546 pKa = 11.84SGYY549 pKa = 8.22GQPIASTLSNITLPMQDD566 pKa = 3.7NNTDD570 pKa = 3.69VYY572 pKa = 10.5CVRR575 pKa = 11.84SNQFSVYY582 pKa = 8.11VHH584 pKa = 5.76STCKK588 pKa = 10.33SSSWDD593 pKa = 3.38NIFNQDD599 pKa = 3.41CTDD602 pKa = 3.44VLEE605 pKa = 4.39ATAVIKK611 pKa = 9.94TGTCPFSFDD620 pKa = 3.91KK621 pKa = 11.18LNNYY625 pKa = 7.76LTFNKK630 pKa = 9.63FCLSLSPVGANCKK643 pKa = 9.97FDD645 pKa = 3.74VAAHH649 pKa = 5.81TRR651 pKa = 11.84TNEE654 pKa = 3.69QVVRR658 pKa = 11.84SLYY661 pKa = 10.45VIYY664 pKa = 10.55EE665 pKa = 4.03EE666 pKa = 4.75GDD668 pKa = 3.7NIVGVPSDD676 pKa = 3.48NSGLHH681 pKa = 6.34DD682 pKa = 5.3LSVLHH687 pKa = 7.02LDD689 pKa = 3.23SCTDD693 pKa = 3.23YY694 pKa = 11.33NIYY697 pKa = 10.59GRR699 pKa = 11.84TGVGIIRR706 pKa = 11.84QTNSTLLSGLYY717 pKa = 8.14YY718 pKa = 10.47TSLSGDD724 pKa = 3.35LLGFKK729 pKa = 10.1NVSDD733 pKa = 3.82GVIYY737 pKa = 10.67SVTPCDD743 pKa = 3.31VSAQVAVIDD752 pKa = 4.2GAIVGAMTSINSEE765 pKa = 4.32LLGLTHH771 pKa = 7.13WTTTPNFYY779 pKa = 10.7YY780 pKa = 10.66YY781 pKa = 10.66SIYY784 pKa = 10.78NYY786 pKa = 9.49TNEE789 pKa = 4.22RR790 pKa = 11.84TRR792 pKa = 11.84GTAIDD797 pKa = 4.46SNDD800 pKa = 3.38VDD802 pKa = 4.54CEE804 pKa = 4.34PIITYY809 pKa = 11.09ANIGVCKK816 pKa = 10.5NGALVFINVTHH827 pKa = 7.03SDD829 pKa = 3.31GDD831 pKa = 3.76VQPISTGNVTIPTNFTISVQVEE853 pKa = 4.08YY854 pKa = 10.31IQVYY858 pKa = 5.57TTPVSIDD865 pKa = 3.21CSRR868 pKa = 11.84YY869 pKa = 8.47VCNGNPRR876 pKa = 11.84CNKK879 pKa = 10.11LLTQYY884 pKa = 10.84VSACQTIEE892 pKa = 3.6QALAMGARR900 pKa = 11.84LEE902 pKa = 4.0NMEE905 pKa = 4.38VDD907 pKa = 3.28SMLFVSEE914 pKa = 4.35NALKK918 pKa = 10.36LASVEE923 pKa = 4.2AFNSTEE929 pKa = 3.91NLDD932 pKa = 4.31PIYY935 pKa = 10.67KK936 pKa = 9.41EE937 pKa = 3.84WPNIGGSWLGGLKK950 pKa = 10.5DD951 pKa = 4.17ILPSHH956 pKa = 5.68NSKK959 pKa = 10.42RR960 pKa = 11.84KK961 pKa = 8.03YY962 pKa = 10.0RR963 pKa = 11.84SAIEE967 pKa = 4.79DD968 pKa = 3.77LLFDD972 pKa = 3.96KK973 pKa = 11.1VVTSGLGTVDD983 pKa = 3.79EE984 pKa = 4.94DD985 pKa = 4.2YY986 pKa = 11.18KK987 pKa = 11.08RR988 pKa = 11.84CTGGYY993 pKa = 10.17DD994 pKa = 3.45IADD997 pKa = 4.27LVCAQYY1003 pKa = 11.58YY1004 pKa = 8.99NGIMVLPGVANDD1016 pKa = 5.23DD1017 pKa = 3.64KK1018 pKa = 10.09MTMYY1022 pKa = 9.25TASLAGGITLGALGGGAVAIPFAVAVQARR1051 pKa = 11.84LNYY1054 pKa = 9.72VALQTDD1060 pKa = 4.12VLNKK1064 pKa = 9.83NQQILANAFNQAIGNITQAFGKK1086 pKa = 10.65VNDD1089 pKa = 5.6AIHH1092 pKa = 5.36QTSQGLATVAKK1103 pKa = 10.13ALAKK1107 pKa = 10.36VQDD1110 pKa = 3.83VVNTQGQALSHH1121 pKa = 6.03LTVQLQNNFQAISSSISDD1139 pKa = 3.33IYY1141 pKa = 11.39NRR1143 pKa = 11.84LDD1145 pKa = 3.61EE1146 pKa = 4.93LSADD1150 pKa = 3.68AQVDD1154 pKa = 3.66RR1155 pKa = 11.84LITGRR1160 pKa = 11.84LTALNAFVSQTLTRR1174 pKa = 11.84QAEE1177 pKa = 4.39VRR1179 pKa = 11.84ASRR1182 pKa = 11.84QLAKK1186 pKa = 10.86DD1187 pKa = 3.4KK1188 pKa = 11.38VNEE1191 pKa = 4.27CVRR1194 pKa = 11.84SQSQRR1199 pKa = 11.84FGFCGNGTHH1208 pKa = 7.49LFSLANAAPNGMIFFHH1224 pKa = 6.14TVLLPTAYY1232 pKa = 8.01EE1233 pKa = 4.33TVTAWSGICASDD1245 pKa = 3.56GDD1247 pKa = 4.1RR1248 pKa = 11.84TFGLVVKK1255 pKa = 10.13DD1256 pKa = 3.57VQLTLFRR1263 pKa = 11.84NLDD1266 pKa = 3.49GKK1268 pKa = 11.0FYY1270 pKa = 9.4LTPRR1274 pKa = 11.84TMYY1277 pKa = 9.68QPRR1280 pKa = 11.84VVTSSDD1286 pKa = 3.09FVQIEE1291 pKa = 4.46GCDD1294 pKa = 3.54VLFVNATVIDD1304 pKa = 4.59LPSIIPDD1311 pKa = 3.84YY1312 pKa = 11.09IDD1314 pKa = 3.75INQTVQDD1321 pKa = 3.37ILEE1324 pKa = 4.31NYY1326 pKa = 8.5RR1327 pKa = 11.84PNWTVPEE1334 pKa = 4.11LTLDD1338 pKa = 3.76IFNATYY1344 pKa = 11.19LNLTGEE1350 pKa = 4.13IDD1352 pKa = 3.65DD1353 pKa = 5.06LEE1355 pKa = 4.33FRR1357 pKa = 11.84SEE1359 pKa = 4.1KK1360 pKa = 10.53LHH1362 pKa = 5.34NTTVEE1367 pKa = 3.84LAILIDD1373 pKa = 4.43NINNTLVNLEE1383 pKa = 3.92WLNRR1387 pKa = 11.84IEE1389 pKa = 5.6TYY1391 pKa = 10.75VKK1393 pKa = 9.1WPWYY1397 pKa = 8.39VWLLIGLVVIFCIPLLLFCCCSTGCCGCIGCLGSCCHH1434 pKa = 6.85SICSRR1439 pKa = 11.84RR1440 pKa = 11.84QFEE1443 pKa = 4.2NYY1445 pKa = 9.36EE1446 pKa = 4.38PIEE1449 pKa = 4.26KK1450 pKa = 10.17VHH1452 pKa = 5.03VHH1454 pKa = 6.07
Molecular weight: 161.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9TEX3|H9TEX3_9ALPC 3C OS=Canine coronavirus OX=11153 PE=4 SV=1
MM1 pKa = 7.51LSLVSPLLKK10 pKa = 10.35KK11 pKa = 10.86SIVIQLFNITVYY23 pKa = 10.74KK24 pKa = 10.02FQAKK28 pKa = 8.76FWYY31 pKa = 9.49KK32 pKa = 10.81LPFEE36 pKa = 4.4TRR38 pKa = 11.84LCIIKK43 pKa = 8.63HH44 pKa = 5.46TKK46 pKa = 9.52PKK48 pKa = 10.45ALSATKK54 pKa = 9.69QVKK57 pKa = 9.41RR58 pKa = 11.84DD59 pKa = 3.56YY60 pKa = 10.98RR61 pKa = 11.84KK62 pKa = 9.79AAILNSMRR70 pKa = 11.84KK71 pKa = 9.08
MM1 pKa = 7.51LSLVSPLLKK10 pKa = 10.35KK11 pKa = 10.86SIVIQLFNITVYY23 pKa = 10.74KK24 pKa = 10.02FQAKK28 pKa = 8.76FWYY31 pKa = 9.49KK32 pKa = 10.81LPFEE36 pKa = 4.4TRR38 pKa = 11.84LCIIKK43 pKa = 8.63HH44 pKa = 5.46TKK46 pKa = 9.52PKK48 pKa = 10.45ALSATKK54 pKa = 9.69QVKK57 pKa = 9.41RR58 pKa = 11.84DD59 pKa = 3.56YY60 pKa = 10.98RR61 pKa = 11.84KK62 pKa = 9.79AAILNSMRR70 pKa = 11.84KK71 pKa = 9.08
Molecular weight: 8.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13608 |
71 |
6685 |
1237.1 |
138.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.481 ± 0.177 | 3.094 ± 0.282 |
5.409 ± 0.252 | 5.049 ± 0.398 |
5.489 ± 0.18 | 6.32 ± 0.145 |
1.808 ± 0.173 | 5.989 ± 0.268 |
6.43 ± 0.808 | 8.392 ± 0.526 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.337 ± 0.304 | 5.938 ± 0.442 |
3.241 ± 0.1 | 3.057 ± 0.377 |
3.123 ± 0.47 | 6.922 ± 0.267 |
5.938 ± 0.552 | 9.053 ± 0.388 |
1.161 ± 0.113 | 4.769 ± 0.188 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
