
Porcine stool-associated circular virus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
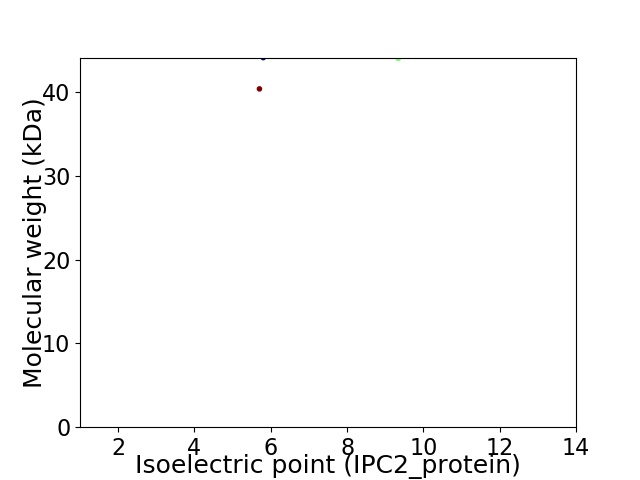
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8TKM5|W8TKM5_9CIRC Replication-associated protein OS=Porcine stool-associated circular virus 4 OX=1475062 PE=4 SV=1
MM1 pKa = 7.86PARR4 pKa = 11.84NPEE7 pKa = 3.69GRR9 pKa = 11.84PQRR12 pKa = 11.84FRR14 pKa = 11.84FACITAWNMDD24 pKa = 4.04AFDD27 pKa = 5.42LDD29 pKa = 3.74WPQRR33 pKa = 11.84MHH35 pKa = 7.46DD36 pKa = 3.95LGLSYY41 pKa = 10.97VVIGRR46 pKa = 11.84EE47 pKa = 3.86TCPRR51 pKa = 11.84TGRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 4.92LQCYY60 pKa = 9.5FEE62 pKa = 4.37ATNPKK67 pKa = 8.86TVTAWQDD74 pKa = 3.18VVMDD78 pKa = 4.11KK79 pKa = 10.78PRR81 pKa = 11.84AHH83 pKa = 6.16VEE85 pKa = 3.9CRR87 pKa = 11.84AAWKK91 pKa = 10.15PEE93 pKa = 3.79GEE95 pKa = 5.2DD96 pKa = 3.11IAEE99 pKa = 4.19NQGDD103 pKa = 4.02KK104 pKa = 11.14AADD107 pKa = 3.59YY108 pKa = 10.18CKK110 pKa = 10.65KK111 pKa = 10.3EE112 pKa = 3.8GDD114 pKa = 3.72FIEE117 pKa = 4.82YY118 pKa = 7.81GTLAPGQGARR128 pKa = 11.84MDD130 pKa = 4.16LADD133 pKa = 4.95CKK135 pKa = 10.88TMIDD139 pKa = 4.09EE140 pKa = 4.9GKK142 pKa = 10.74QMIDD146 pKa = 3.16LYY148 pKa = 9.97EE149 pKa = 4.1KK150 pKa = 10.93HH151 pKa = 6.54FGTCVRR157 pKa = 11.84CHH159 pKa = 6.86RR160 pKa = 11.84GLMLYY165 pKa = 10.28KK166 pKa = 10.64DD167 pKa = 3.46LVDD170 pKa = 3.82RR171 pKa = 11.84KK172 pKa = 9.57RR173 pKa = 11.84RR174 pKa = 11.84KK175 pKa = 8.6EE176 pKa = 3.8AEE178 pKa = 4.2PEE180 pKa = 3.74PKK182 pKa = 10.24EE183 pKa = 3.51IVVYY187 pKa = 10.48VGASGSGKK195 pKa = 8.84SHH197 pKa = 6.67HH198 pKa = 6.92CWHH201 pKa = 7.23DD202 pKa = 3.02PDD204 pKa = 4.03YY205 pKa = 11.15RR206 pKa = 11.84RR207 pKa = 11.84SGYY210 pKa = 10.28KK211 pKa = 10.07YY212 pKa = 10.21PLLAEE217 pKa = 4.11NKK219 pKa = 8.11VWFDD223 pKa = 3.42GYY225 pKa = 10.87EE226 pKa = 4.11GEE228 pKa = 4.22EE229 pKa = 4.12VLWVDD234 pKa = 3.79EE235 pKa = 4.38FRR237 pKa = 11.84GSVFPFGLFLQVTDD251 pKa = 2.78KK252 pKa = 10.38WGARR256 pKa = 11.84VEE258 pKa = 4.36VKK260 pKa = 10.49GGSIEE265 pKa = 4.25TFFKK269 pKa = 10.73KK270 pKa = 10.62VLISTTVPPGEE281 pKa = 4.13WYY283 pKa = 10.48KK284 pKa = 11.27NCSNYY289 pKa = 10.47LKK291 pKa = 10.52NPKK294 pKa = 9.09QLWRR298 pKa = 11.84RR299 pKa = 11.84LTKK302 pKa = 10.28VYY304 pKa = 10.1WLGPVEE310 pKa = 4.03QDD312 pKa = 3.51DD313 pKa = 4.81DD314 pKa = 4.4GFTIYY319 pKa = 10.24PEE321 pKa = 4.53PEE323 pKa = 5.18LIPDD327 pKa = 4.21PEE329 pKa = 5.89HH330 pKa = 7.0DD331 pKa = 3.29TRR333 pKa = 11.84TNPFYY338 pKa = 11.11DD339 pKa = 3.3GVGTHH344 pKa = 5.99YY345 pKa = 10.45PRR347 pKa = 11.84EE348 pKa = 4.12
MM1 pKa = 7.86PARR4 pKa = 11.84NPEE7 pKa = 3.69GRR9 pKa = 11.84PQRR12 pKa = 11.84FRR14 pKa = 11.84FACITAWNMDD24 pKa = 4.04AFDD27 pKa = 5.42LDD29 pKa = 3.74WPQRR33 pKa = 11.84MHH35 pKa = 7.46DD36 pKa = 3.95LGLSYY41 pKa = 10.97VVIGRR46 pKa = 11.84EE47 pKa = 3.86TCPRR51 pKa = 11.84TGRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 4.92LQCYY60 pKa = 9.5FEE62 pKa = 4.37ATNPKK67 pKa = 8.86TVTAWQDD74 pKa = 3.18VVMDD78 pKa = 4.11KK79 pKa = 10.78PRR81 pKa = 11.84AHH83 pKa = 6.16VEE85 pKa = 3.9CRR87 pKa = 11.84AAWKK91 pKa = 10.15PEE93 pKa = 3.79GEE95 pKa = 5.2DD96 pKa = 3.11IAEE99 pKa = 4.19NQGDD103 pKa = 4.02KK104 pKa = 11.14AADD107 pKa = 3.59YY108 pKa = 10.18CKK110 pKa = 10.65KK111 pKa = 10.3EE112 pKa = 3.8GDD114 pKa = 3.72FIEE117 pKa = 4.82YY118 pKa = 7.81GTLAPGQGARR128 pKa = 11.84MDD130 pKa = 4.16LADD133 pKa = 4.95CKK135 pKa = 10.88TMIDD139 pKa = 4.09EE140 pKa = 4.9GKK142 pKa = 10.74QMIDD146 pKa = 3.16LYY148 pKa = 9.97EE149 pKa = 4.1KK150 pKa = 10.93HH151 pKa = 6.54FGTCVRR157 pKa = 11.84CHH159 pKa = 6.86RR160 pKa = 11.84GLMLYY165 pKa = 10.28KK166 pKa = 10.64DD167 pKa = 3.46LVDD170 pKa = 3.82RR171 pKa = 11.84KK172 pKa = 9.57RR173 pKa = 11.84RR174 pKa = 11.84KK175 pKa = 8.6EE176 pKa = 3.8AEE178 pKa = 4.2PEE180 pKa = 3.74PKK182 pKa = 10.24EE183 pKa = 3.51IVVYY187 pKa = 10.48VGASGSGKK195 pKa = 8.84SHH197 pKa = 6.67HH198 pKa = 6.92CWHH201 pKa = 7.23DD202 pKa = 3.02PDD204 pKa = 4.03YY205 pKa = 11.15RR206 pKa = 11.84RR207 pKa = 11.84SGYY210 pKa = 10.28KK211 pKa = 10.07YY212 pKa = 10.21PLLAEE217 pKa = 4.11NKK219 pKa = 8.11VWFDD223 pKa = 3.42GYY225 pKa = 10.87EE226 pKa = 4.11GEE228 pKa = 4.22EE229 pKa = 4.12VLWVDD234 pKa = 3.79EE235 pKa = 4.38FRR237 pKa = 11.84GSVFPFGLFLQVTDD251 pKa = 2.78KK252 pKa = 10.38WGARR256 pKa = 11.84VEE258 pKa = 4.36VKK260 pKa = 10.49GGSIEE265 pKa = 4.25TFFKK269 pKa = 10.73KK270 pKa = 10.62VLISTTVPPGEE281 pKa = 4.13WYY283 pKa = 10.48KK284 pKa = 11.27NCSNYY289 pKa = 10.47LKK291 pKa = 10.52NPKK294 pKa = 9.09QLWRR298 pKa = 11.84RR299 pKa = 11.84LTKK302 pKa = 10.28VYY304 pKa = 10.1WLGPVEE310 pKa = 4.03QDD312 pKa = 3.51DD313 pKa = 4.81DD314 pKa = 4.4GFTIYY319 pKa = 10.24PEE321 pKa = 4.53PEE323 pKa = 5.18LIPDD327 pKa = 4.21PEE329 pKa = 5.89HH330 pKa = 7.0DD331 pKa = 3.29TRR333 pKa = 11.84TNPFYY338 pKa = 11.11DD339 pKa = 3.3GVGTHH344 pKa = 5.99YY345 pKa = 10.45PRR347 pKa = 11.84EE348 pKa = 4.12
Molecular weight: 40.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8TKM5|W8TKM5_9CIRC Replication-associated protein OS=Porcine stool-associated circular virus 4 OX=1475062 PE=4 SV=1
MM1 pKa = 7.26HH2 pKa = 7.81RR3 pKa = 11.84SGDD6 pKa = 3.64LALFLYY12 pKa = 9.97VVVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.05RR20 pKa = 11.84KK21 pKa = 9.39SRR23 pKa = 11.84GRR25 pKa = 11.84LLRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84ARR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 9.62RR35 pKa = 11.84LSKK38 pKa = 9.65SMRR41 pKa = 11.84MRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84TIRR50 pKa = 11.84RR51 pKa = 11.84NRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84GGLTVSGSAEE65 pKa = 3.94YY66 pKa = 9.46PLSYY70 pKa = 10.96GSLLPVEE77 pKa = 5.45RR78 pKa = 11.84KK79 pKa = 10.1AKK81 pKa = 10.21GSDD84 pKa = 2.87VGVPHH89 pKa = 7.62DD90 pKa = 5.38GFCTWSICPLPMGVPQVIGDD110 pKa = 3.97LSVVSFWADD119 pKa = 3.06LNTSYY124 pKa = 11.7GEE126 pKa = 4.08RR127 pKa = 11.84YY128 pKa = 8.92KK129 pKa = 10.95PKK131 pKa = 10.24IGLGQNASDD140 pKa = 3.96EE141 pKa = 4.57TKK143 pKa = 9.88MLCDD147 pKa = 4.51LPVCLSSFTVRR158 pKa = 11.84SIVKK162 pKa = 10.5CEE164 pKa = 4.1TLWKK168 pKa = 10.14LYY170 pKa = 10.57KK171 pKa = 10.25LYY173 pKa = 10.4RR174 pKa = 11.84ISWVSVTFTVPEE186 pKa = 4.0FTGGQRR192 pKa = 11.84NHH194 pKa = 6.08NLYY197 pKa = 10.87LEE199 pKa = 4.41WTHH202 pKa = 6.87LPKK205 pKa = 10.48ARR207 pKa = 11.84CAAYY211 pKa = 9.25EE212 pKa = 4.0DD213 pKa = 4.83CIGMVVSGNGKK224 pKa = 10.16SSDD227 pKa = 3.33TGGWNWIYY235 pKa = 10.88NPPDD239 pKa = 3.13VATACSIDD247 pKa = 3.16GRR249 pKa = 11.84KK250 pKa = 9.61NGRR253 pKa = 11.84NGWRR257 pKa = 11.84RR258 pKa = 11.84AQLAYY263 pKa = 10.23NHH265 pKa = 7.09PVTISWRR272 pKa = 11.84PKK274 pKa = 9.17HH275 pKa = 6.65AEE277 pKa = 4.12LIQDD281 pKa = 3.74HH282 pKa = 5.97VNYY285 pKa = 9.81WDD287 pKa = 3.55ATSASTGGTKK297 pKa = 10.51VMDD300 pKa = 3.93LWPTKK305 pKa = 10.74DD306 pKa = 3.16KK307 pKa = 10.74FVRR310 pKa = 11.84SYY312 pKa = 11.34LPTDD316 pKa = 3.67IDD318 pKa = 3.53QQMTAEE324 pKa = 4.08RR325 pKa = 11.84QVWCGPVIRR334 pKa = 11.84LIDD337 pKa = 3.9ADD339 pKa = 3.52ITAAAIPSQPPTNTWYY355 pKa = 9.25SQYY358 pKa = 10.98GIRR361 pKa = 11.84CTCVVKK367 pKa = 10.83LKK369 pKa = 10.17FRR371 pKa = 11.84NMDD374 pKa = 3.45AADD377 pKa = 4.77PIFPEE382 pKa = 4.39YY383 pKa = 11.0VPP385 pKa = 4.56
MM1 pKa = 7.26HH2 pKa = 7.81RR3 pKa = 11.84SGDD6 pKa = 3.64LALFLYY12 pKa = 9.97VVVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.05RR20 pKa = 11.84KK21 pKa = 9.39SRR23 pKa = 11.84GRR25 pKa = 11.84LLRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84ARR32 pKa = 11.84RR33 pKa = 11.84YY34 pKa = 9.62RR35 pKa = 11.84LSKK38 pKa = 9.65SMRR41 pKa = 11.84MRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84TIRR50 pKa = 11.84RR51 pKa = 11.84NRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84GGLTVSGSAEE65 pKa = 3.94YY66 pKa = 9.46PLSYY70 pKa = 10.96GSLLPVEE77 pKa = 5.45RR78 pKa = 11.84KK79 pKa = 10.1AKK81 pKa = 10.21GSDD84 pKa = 2.87VGVPHH89 pKa = 7.62DD90 pKa = 5.38GFCTWSICPLPMGVPQVIGDD110 pKa = 3.97LSVVSFWADD119 pKa = 3.06LNTSYY124 pKa = 11.7GEE126 pKa = 4.08RR127 pKa = 11.84YY128 pKa = 8.92KK129 pKa = 10.95PKK131 pKa = 10.24IGLGQNASDD140 pKa = 3.96EE141 pKa = 4.57TKK143 pKa = 9.88MLCDD147 pKa = 4.51LPVCLSSFTVRR158 pKa = 11.84SIVKK162 pKa = 10.5CEE164 pKa = 4.1TLWKK168 pKa = 10.14LYY170 pKa = 10.57KK171 pKa = 10.25LYY173 pKa = 10.4RR174 pKa = 11.84ISWVSVTFTVPEE186 pKa = 4.0FTGGQRR192 pKa = 11.84NHH194 pKa = 6.08NLYY197 pKa = 10.87LEE199 pKa = 4.41WTHH202 pKa = 6.87LPKK205 pKa = 10.48ARR207 pKa = 11.84CAAYY211 pKa = 9.25EE212 pKa = 4.0DD213 pKa = 4.83CIGMVVSGNGKK224 pKa = 10.16SSDD227 pKa = 3.33TGGWNWIYY235 pKa = 10.88NPPDD239 pKa = 3.13VATACSIDD247 pKa = 3.16GRR249 pKa = 11.84KK250 pKa = 9.61NGRR253 pKa = 11.84NGWRR257 pKa = 11.84RR258 pKa = 11.84AQLAYY263 pKa = 10.23NHH265 pKa = 7.09PVTISWRR272 pKa = 11.84PKK274 pKa = 9.17HH275 pKa = 6.65AEE277 pKa = 4.12LIQDD281 pKa = 3.74HH282 pKa = 5.97VNYY285 pKa = 9.81WDD287 pKa = 3.55ATSASTGGTKK297 pKa = 10.51VMDD300 pKa = 3.93LWPTKK305 pKa = 10.74DD306 pKa = 3.16KK307 pKa = 10.74FVRR310 pKa = 11.84SYY312 pKa = 11.34LPTDD316 pKa = 3.67IDD318 pKa = 3.53QQMTAEE324 pKa = 4.08RR325 pKa = 11.84QVWCGPVIRR334 pKa = 11.84LIDD337 pKa = 3.9ADD339 pKa = 3.52ITAAAIPSQPPTNTWYY355 pKa = 9.25SQYY358 pKa = 10.98GIRR361 pKa = 11.84CTCVVKK367 pKa = 10.83LKK369 pKa = 10.17FRR371 pKa = 11.84NMDD374 pKa = 3.45AADD377 pKa = 4.77PIFPEE382 pKa = 4.39YY383 pKa = 11.0VPP385 pKa = 4.56
Molecular weight: 43.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
733 |
348 |
385 |
366.5 |
42.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.73 ± 0.168 | 2.865 ± 0.005 |
6.548 ± 0.754 | 5.321 ± 1.697 |
3.274 ± 0.645 | 7.776 ± 0.347 |
2.319 ± 0.345 | 3.956 ± 0.495 |
5.866 ± 0.642 | 6.821 ± 0.49 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.319 ± 0.013 | 3.138 ± 0.344 |
6.412 ± 0.302 | 2.729 ± 0.09 |
8.868 ± 1.049 | 5.184 ± 1.618 |
5.73 ± 0.347 | 7.094 ± 0.302 |
3.274 ± 0.071 | 4.775 ± 0.069 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
