
Human parainfluenza 2 virus (strain Toshiba) (HPIV-2)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Orthorubulavirus; Human orthorubulavirus 2
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
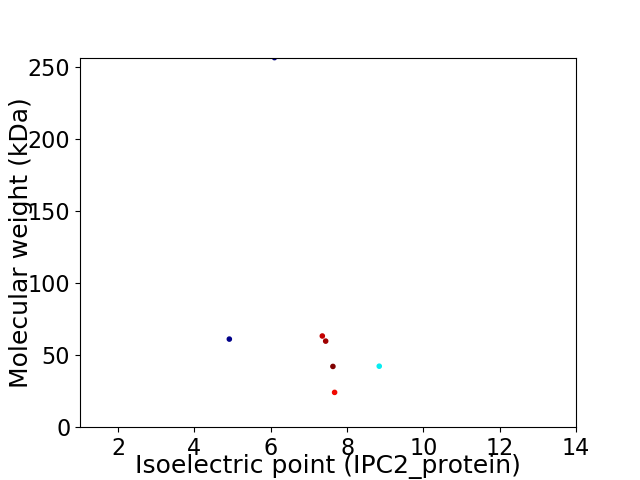
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P23056|PHOSP_PI2HT Phosphoprotein OS=Human parainfluenza 2 virus (strain Toshiba) OX=11214 GN=P/V PE=3 SV=1
MM1 pKa = 7.45SSVLKK6 pKa = 9.72TFEE9 pKa = 4.27RR10 pKa = 11.84FTIQQEE16 pKa = 4.18LQEE19 pKa = 4.44QSDD22 pKa = 4.1DD23 pKa = 3.83TPVPLEE29 pKa = 4.27TIKK32 pKa = 9.66PTIRR36 pKa = 11.84VFVINNNDD44 pKa = 3.28PVVRR48 pKa = 11.84SRR50 pKa = 11.84LLFFNLRR57 pKa = 11.84IIMSNTARR65 pKa = 11.84EE66 pKa = 4.14GHH68 pKa = 6.37RR69 pKa = 11.84AGALLSLLSLPSAAMSNHH87 pKa = 6.13IKK89 pKa = 10.55LAMHH93 pKa = 6.37SPEE96 pKa = 3.82ASIDD100 pKa = 3.53RR101 pKa = 11.84VEE103 pKa = 3.97ITGFEE108 pKa = 4.17NNSFRR113 pKa = 11.84VIPDD117 pKa = 2.97ARR119 pKa = 11.84STMSRR124 pKa = 11.84GEE126 pKa = 3.95VLAFEE131 pKa = 5.18ALAEE135 pKa = 5.23DD136 pKa = 4.85IPDD139 pKa = 4.02TLNHH143 pKa = 5.17QTPFVNNDD151 pKa = 2.92VEE153 pKa = 5.65DD154 pKa = 5.53DD155 pKa = 3.81IFDD158 pKa = 3.73EE159 pKa = 4.62TEE161 pKa = 3.79KK162 pKa = 10.98FLDD165 pKa = 3.29VCYY168 pKa = 10.53SVLMQAWIVTCKK180 pKa = 10.52CMTAPDD186 pKa = 3.93QPPVSVAKK194 pKa = 8.98MAKK197 pKa = 8.5YY198 pKa = 9.12QQQGRR203 pKa = 11.84INARR207 pKa = 11.84YY208 pKa = 8.82VLQPEE213 pKa = 4.31AQRR216 pKa = 11.84LIQNAIRR223 pKa = 11.84KK224 pKa = 9.25SMVVRR229 pKa = 11.84HH230 pKa = 5.66FMTYY234 pKa = 9.61EE235 pKa = 3.85LQLSQSRR242 pKa = 11.84SLLANRR248 pKa = 11.84YY249 pKa = 8.02YY250 pKa = 11.74AMVGDD255 pKa = 3.33IGKK258 pKa = 9.94YY259 pKa = 9.26IEE261 pKa = 4.21HH262 pKa = 6.88SGMGGFFLTLKK273 pKa = 10.42YY274 pKa = 10.88GLGTRR279 pKa = 11.84WPTLALAAFSGEE291 pKa = 4.06LQKK294 pKa = 11.25LKK296 pKa = 11.14ALMLHH301 pKa = 5.45YY302 pKa = 10.47QSLGPMAKK310 pKa = 10.14YY311 pKa = 8.52MALLEE316 pKa = 4.47SPKK319 pKa = 10.97LMDD322 pKa = 4.6FVPSEE327 pKa = 4.17YY328 pKa = 10.32PLDD331 pKa = 3.47YY332 pKa = 10.53SYY334 pKa = 12.07AMGIGTVLDD343 pKa = 3.74TNMRR347 pKa = 11.84NYY349 pKa = 10.98AYY351 pKa = 10.24GRR353 pKa = 11.84SYY355 pKa = 11.47LNQQYY360 pKa = 8.84FQLGVEE366 pKa = 4.63TARR369 pKa = 11.84KK370 pKa = 7.79QQGAVDD376 pKa = 3.5NRR378 pKa = 11.84TAEE381 pKa = 4.24DD382 pKa = 3.74LGMTAADD389 pKa = 4.18KK390 pKa = 11.39ADD392 pKa = 3.54LTATISKK399 pKa = 10.37LSLSQLPRR407 pKa = 11.84GRR409 pKa = 11.84QPISDD414 pKa = 4.38PFAGANDD421 pKa = 3.87RR422 pKa = 11.84EE423 pKa = 4.54MGGQANDD430 pKa = 3.53TPVYY434 pKa = 10.22NFNPIDD440 pKa = 3.35TRR442 pKa = 11.84RR443 pKa = 11.84YY444 pKa = 10.7DD445 pKa = 3.68NYY447 pKa = 11.32DD448 pKa = 2.77SDD450 pKa = 4.84GEE452 pKa = 4.09DD453 pKa = 5.21RR454 pKa = 11.84IDD456 pKa = 3.8NDD458 pKa = 2.44QDD460 pKa = 3.19QAIRR464 pKa = 11.84EE465 pKa = 4.17NRR467 pKa = 11.84GEE469 pKa = 3.96PGQPNNQTSDD479 pKa = 2.88NQQRR483 pKa = 11.84FNPPIPQRR491 pKa = 11.84TSGMSSEE498 pKa = 4.47EE499 pKa = 4.09FQHH502 pKa = 7.27SMNQYY507 pKa = 8.22IRR509 pKa = 11.84AMHH512 pKa = 5.25EE513 pKa = 4.07QYY515 pKa = 10.5RR516 pKa = 11.84GSQDD520 pKa = 4.47DD521 pKa = 4.19DD522 pKa = 5.77ANDD525 pKa = 3.63ATDD528 pKa = 4.3GNDD531 pKa = 2.89ISLEE535 pKa = 4.02LVGDD539 pKa = 4.32FDD541 pKa = 4.53SS542 pKa = 4.56
MM1 pKa = 7.45SSVLKK6 pKa = 9.72TFEE9 pKa = 4.27RR10 pKa = 11.84FTIQQEE16 pKa = 4.18LQEE19 pKa = 4.44QSDD22 pKa = 4.1DD23 pKa = 3.83TPVPLEE29 pKa = 4.27TIKK32 pKa = 9.66PTIRR36 pKa = 11.84VFVINNNDD44 pKa = 3.28PVVRR48 pKa = 11.84SRR50 pKa = 11.84LLFFNLRR57 pKa = 11.84IIMSNTARR65 pKa = 11.84EE66 pKa = 4.14GHH68 pKa = 6.37RR69 pKa = 11.84AGALLSLLSLPSAAMSNHH87 pKa = 6.13IKK89 pKa = 10.55LAMHH93 pKa = 6.37SPEE96 pKa = 3.82ASIDD100 pKa = 3.53RR101 pKa = 11.84VEE103 pKa = 3.97ITGFEE108 pKa = 4.17NNSFRR113 pKa = 11.84VIPDD117 pKa = 2.97ARR119 pKa = 11.84STMSRR124 pKa = 11.84GEE126 pKa = 3.95VLAFEE131 pKa = 5.18ALAEE135 pKa = 5.23DD136 pKa = 4.85IPDD139 pKa = 4.02TLNHH143 pKa = 5.17QTPFVNNDD151 pKa = 2.92VEE153 pKa = 5.65DD154 pKa = 5.53DD155 pKa = 3.81IFDD158 pKa = 3.73EE159 pKa = 4.62TEE161 pKa = 3.79KK162 pKa = 10.98FLDD165 pKa = 3.29VCYY168 pKa = 10.53SVLMQAWIVTCKK180 pKa = 10.52CMTAPDD186 pKa = 3.93QPPVSVAKK194 pKa = 8.98MAKK197 pKa = 8.5YY198 pKa = 9.12QQQGRR203 pKa = 11.84INARR207 pKa = 11.84YY208 pKa = 8.82VLQPEE213 pKa = 4.31AQRR216 pKa = 11.84LIQNAIRR223 pKa = 11.84KK224 pKa = 9.25SMVVRR229 pKa = 11.84HH230 pKa = 5.66FMTYY234 pKa = 9.61EE235 pKa = 3.85LQLSQSRR242 pKa = 11.84SLLANRR248 pKa = 11.84YY249 pKa = 8.02YY250 pKa = 11.74AMVGDD255 pKa = 3.33IGKK258 pKa = 9.94YY259 pKa = 9.26IEE261 pKa = 4.21HH262 pKa = 6.88SGMGGFFLTLKK273 pKa = 10.42YY274 pKa = 10.88GLGTRR279 pKa = 11.84WPTLALAAFSGEE291 pKa = 4.06LQKK294 pKa = 11.25LKK296 pKa = 11.14ALMLHH301 pKa = 5.45YY302 pKa = 10.47QSLGPMAKK310 pKa = 10.14YY311 pKa = 8.52MALLEE316 pKa = 4.47SPKK319 pKa = 10.97LMDD322 pKa = 4.6FVPSEE327 pKa = 4.17YY328 pKa = 10.32PLDD331 pKa = 3.47YY332 pKa = 10.53SYY334 pKa = 12.07AMGIGTVLDD343 pKa = 3.74TNMRR347 pKa = 11.84NYY349 pKa = 10.98AYY351 pKa = 10.24GRR353 pKa = 11.84SYY355 pKa = 11.47LNQQYY360 pKa = 8.84FQLGVEE366 pKa = 4.63TARR369 pKa = 11.84KK370 pKa = 7.79QQGAVDD376 pKa = 3.5NRR378 pKa = 11.84TAEE381 pKa = 4.24DD382 pKa = 3.74LGMTAADD389 pKa = 4.18KK390 pKa = 11.39ADD392 pKa = 3.54LTATISKK399 pKa = 10.37LSLSQLPRR407 pKa = 11.84GRR409 pKa = 11.84QPISDD414 pKa = 4.38PFAGANDD421 pKa = 3.87RR422 pKa = 11.84EE423 pKa = 4.54MGGQANDD430 pKa = 3.53TPVYY434 pKa = 10.22NFNPIDD440 pKa = 3.35TRR442 pKa = 11.84RR443 pKa = 11.84YY444 pKa = 10.7DD445 pKa = 3.68NYY447 pKa = 11.32DD448 pKa = 2.77SDD450 pKa = 4.84GEE452 pKa = 4.09DD453 pKa = 5.21RR454 pKa = 11.84IDD456 pKa = 3.8NDD458 pKa = 2.44QDD460 pKa = 3.19QAIRR464 pKa = 11.84EE465 pKa = 4.17NRR467 pKa = 11.84GEE469 pKa = 3.96PGQPNNQTSDD479 pKa = 2.88NQQRR483 pKa = 11.84FNPPIPQRR491 pKa = 11.84TSGMSSEE498 pKa = 4.47EE499 pKa = 4.09FQHH502 pKa = 7.27SMNQYY507 pKa = 8.22IRR509 pKa = 11.84AMHH512 pKa = 5.25EE513 pKa = 4.07QYY515 pKa = 10.5RR516 pKa = 11.84GSQDD520 pKa = 4.47DD521 pKa = 4.19DD522 pKa = 5.77ANDD525 pKa = 3.63ATDD528 pKa = 4.3GNDD531 pKa = 2.89ISLEE535 pKa = 4.02LVGDD539 pKa = 4.32FDD541 pKa = 4.53SS542 pKa = 4.56
Molecular weight: 61.12 kDa
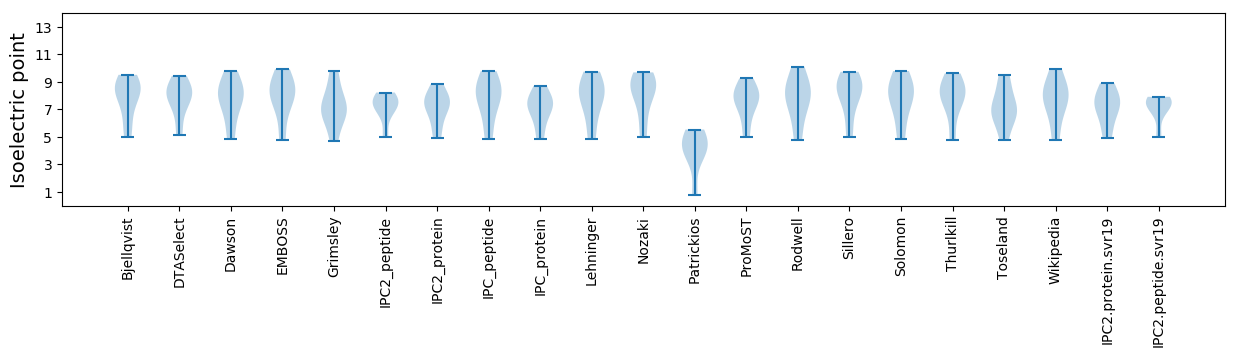
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P25466|HN_PI2HT Hemagglutinin-neuraminidase OS=Human parainfluenza 2 virus (strain Toshiba) OX=11214 GN=HN PE=2 SV=1
MM1 pKa = 7.89PIISLPADD9 pKa = 3.61PTSPSQSLTPFPIQLDD25 pKa = 4.04TKK27 pKa = 10.51DD28 pKa = 3.56GKK30 pKa = 10.62AGKK33 pKa = 9.25LLKK36 pKa = 10.12QIRR39 pKa = 11.84IRR41 pKa = 11.84YY42 pKa = 8.65LNEE45 pKa = 3.71PNSRR49 pKa = 11.84HH50 pKa = 5.11TPITFINTYY59 pKa = 9.03GFVYY63 pKa = 10.59ARR65 pKa = 11.84DD66 pKa = 3.65TSGGIHH72 pKa = 6.18SEE74 pKa = 3.78ISSDD78 pKa = 3.47LAAGSITACMMKK90 pKa = 10.34LGPGPNIQNANLVLRR105 pKa = 11.84SLNEE109 pKa = 3.94FYY111 pKa = 11.5VKK113 pKa = 10.25VKK115 pKa = 9.13KK116 pKa = 9.1TSSQRR121 pKa = 11.84EE122 pKa = 3.82EE123 pKa = 4.0AVFEE127 pKa = 4.18LVNIPTLLRR136 pKa = 11.84EE137 pKa = 3.93HH138 pKa = 7.01ALCKK142 pKa = 10.28RR143 pKa = 11.84KK144 pKa = 8.9MLVCSAEE151 pKa = 4.19KK152 pKa = 10.24FLKK155 pKa = 10.77NPSKK159 pKa = 10.58LQAGFEE165 pKa = 4.24YY166 pKa = 10.76VYY168 pKa = 10.54IPTFVSITYY177 pKa = 10.1SPRR180 pKa = 11.84NLNYY184 pKa = 9.83QVARR188 pKa = 11.84PILKK192 pKa = 8.9FRR194 pKa = 11.84SRR196 pKa = 11.84FVYY199 pKa = 10.36SIHH202 pKa = 7.32LEE204 pKa = 4.3LILRR208 pKa = 11.84LLCKK212 pKa = 10.1SDD214 pKa = 3.63SPLMKK219 pKa = 10.11SYY221 pKa = 10.94NADD224 pKa = 3.52RR225 pKa = 11.84TGRR228 pKa = 11.84GCLASVWIHH237 pKa = 5.23VCNILKK243 pKa = 10.41NKK245 pKa = 10.02SIKK248 pKa = 8.63QQGRR252 pKa = 11.84EE253 pKa = 3.84SYY255 pKa = 10.7FIAKK259 pKa = 8.94CMSMQLQVSIADD271 pKa = 3.39LWGPTIIIKK280 pKa = 10.54SLGHH284 pKa = 5.95IPKK287 pKa = 8.96TALPFFSKK295 pKa = 10.69DD296 pKa = 3.19GIACHH301 pKa = 6.54PLQDD305 pKa = 4.33VSPNLAKK312 pKa = 10.67SLWSVGCEE320 pKa = 3.48IEE322 pKa = 4.31SAKK325 pKa = 10.85LILQEE330 pKa = 4.16SDD332 pKa = 3.5LNEE335 pKa = 4.22LMGHH339 pKa = 6.05QDD341 pKa = 5.06LITDD345 pKa = 4.79KK346 pKa = 10.78IAIRR350 pKa = 11.84SGQRR354 pKa = 11.84TFEE357 pKa = 3.96RR358 pKa = 11.84SKK360 pKa = 10.54FSPFKK365 pKa = 10.6KK366 pKa = 10.1YY367 pKa = 11.09ASIPNLEE374 pKa = 4.75AINN377 pKa = 3.66
MM1 pKa = 7.89PIISLPADD9 pKa = 3.61PTSPSQSLTPFPIQLDD25 pKa = 4.04TKK27 pKa = 10.51DD28 pKa = 3.56GKK30 pKa = 10.62AGKK33 pKa = 9.25LLKK36 pKa = 10.12QIRR39 pKa = 11.84IRR41 pKa = 11.84YY42 pKa = 8.65LNEE45 pKa = 3.71PNSRR49 pKa = 11.84HH50 pKa = 5.11TPITFINTYY59 pKa = 9.03GFVYY63 pKa = 10.59ARR65 pKa = 11.84DD66 pKa = 3.65TSGGIHH72 pKa = 6.18SEE74 pKa = 3.78ISSDD78 pKa = 3.47LAAGSITACMMKK90 pKa = 10.34LGPGPNIQNANLVLRR105 pKa = 11.84SLNEE109 pKa = 3.94FYY111 pKa = 11.5VKK113 pKa = 10.25VKK115 pKa = 9.13KK116 pKa = 9.1TSSQRR121 pKa = 11.84EE122 pKa = 3.82EE123 pKa = 4.0AVFEE127 pKa = 4.18LVNIPTLLRR136 pKa = 11.84EE137 pKa = 3.93HH138 pKa = 7.01ALCKK142 pKa = 10.28RR143 pKa = 11.84KK144 pKa = 8.9MLVCSAEE151 pKa = 4.19KK152 pKa = 10.24FLKK155 pKa = 10.77NPSKK159 pKa = 10.58LQAGFEE165 pKa = 4.24YY166 pKa = 10.76VYY168 pKa = 10.54IPTFVSITYY177 pKa = 10.1SPRR180 pKa = 11.84NLNYY184 pKa = 9.83QVARR188 pKa = 11.84PILKK192 pKa = 8.9FRR194 pKa = 11.84SRR196 pKa = 11.84FVYY199 pKa = 10.36SIHH202 pKa = 7.32LEE204 pKa = 4.3LILRR208 pKa = 11.84LLCKK212 pKa = 10.1SDD214 pKa = 3.63SPLMKK219 pKa = 10.11SYY221 pKa = 10.94NADD224 pKa = 3.52RR225 pKa = 11.84TGRR228 pKa = 11.84GCLASVWIHH237 pKa = 5.23VCNILKK243 pKa = 10.41NKK245 pKa = 10.02SIKK248 pKa = 8.63QQGRR252 pKa = 11.84EE253 pKa = 3.84SYY255 pKa = 10.7FIAKK259 pKa = 8.94CMSMQLQVSIADD271 pKa = 3.39LWGPTIIIKK280 pKa = 10.54SLGHH284 pKa = 5.95IPKK287 pKa = 8.96TALPFFSKK295 pKa = 10.69DD296 pKa = 3.19GIACHH301 pKa = 6.54PLQDD305 pKa = 4.33VSPNLAKK312 pKa = 10.67SLWSVGCEE320 pKa = 3.48IEE322 pKa = 4.31SAKK325 pKa = 10.85LILQEE330 pKa = 4.16SDD332 pKa = 3.5LNEE335 pKa = 4.22LMGHH339 pKa = 6.05QDD341 pKa = 5.06LITDD345 pKa = 4.79KK346 pKa = 10.78IAIRR350 pKa = 11.84SGQRR354 pKa = 11.84TFEE357 pKa = 3.96RR358 pKa = 11.84SKK360 pKa = 10.54FSPFKK365 pKa = 10.6KK366 pKa = 10.1YY367 pKa = 11.09ASIPNLEE374 pKa = 4.75AINN377 pKa = 3.66
Molecular weight: 42.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4923 |
225 |
2262 |
703.3 |
78.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.805 ± 0.646 | 1.747 ± 0.296 |
4.997 ± 0.508 | 4.591 ± 0.492 |
3.758 ± 0.252 | 5.2 ± 0.402 |
2.173 ± 0.228 | 8.694 ± 0.652 |
4.916 ± 0.475 | 10.42 ± 0.868 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.269 | 5.342 ± 0.513 |
5.322 ± 0.561 | 3.961 ± 0.371 |
4.672 ± 0.354 | 8.613 ± 0.244 |
6.967 ± 0.437 | 4.977 ± 0.349 |
0.975 ± 0.26 | 3.616 ± 0.456 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
