
Pigeon circovirus (PiCV) (Columbid circovirus)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; Pigeon circovirus
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
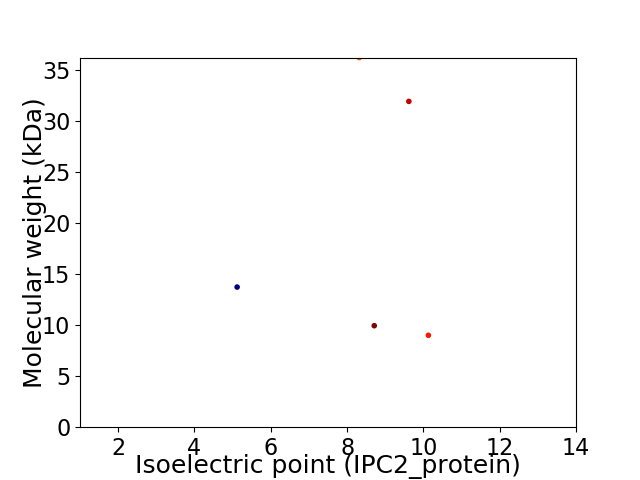
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
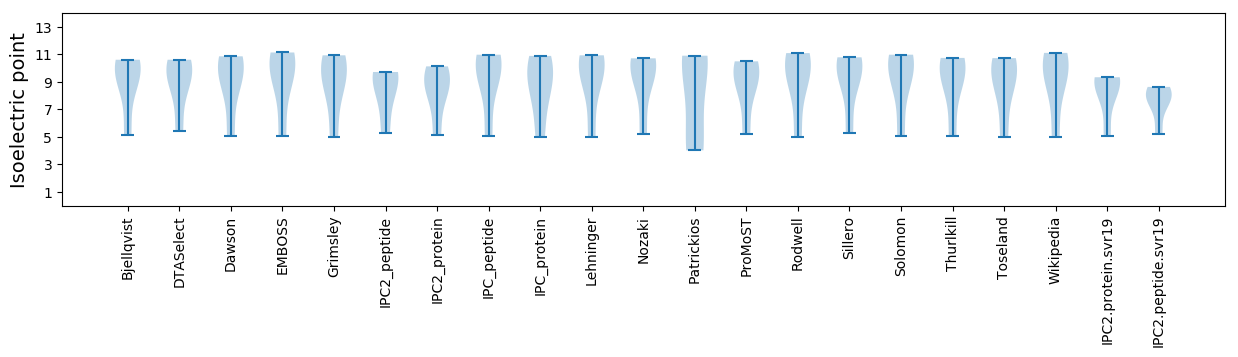
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9IG45|REP_PICV Replication-associated protein OS=Pigeon circovirus OX=126070 GN=rep PE=3 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84VPVGHH7 pKa = 5.75TQQLTEE13 pKa = 4.1GQPAVEE19 pKa = 5.11VINDD23 pKa = 3.6DD24 pKa = 3.69YY25 pKa = 11.47FLVVVPVPPFTLHH38 pKa = 6.49LVEE41 pKa = 4.57FTSGVVSCPTALPATGRR58 pKa = 11.84ARR60 pKa = 11.84DD61 pKa = 3.72DD62 pKa = 3.6DD63 pKa = 4.09DD64 pKa = 4.94FRR66 pKa = 11.84FEE68 pKa = 4.26VTGLLTNQQLQVAQATPIFDD88 pKa = 3.61VYY90 pKa = 9.88LTEE93 pKa = 3.96VSRR96 pKa = 11.84DD97 pKa = 3.47LGHH100 pKa = 6.94CSSGFHH106 pKa = 6.87RR107 pKa = 11.84GNSLAEE113 pKa = 3.84IAPVSLRR120 pKa = 11.84RR121 pKa = 11.84NAQGKK126 pKa = 8.97
MM1 pKa = 7.71RR2 pKa = 11.84VPVGHH7 pKa = 5.75TQQLTEE13 pKa = 4.1GQPAVEE19 pKa = 5.11VINDD23 pKa = 3.6DD24 pKa = 3.69YY25 pKa = 11.47FLVVVPVPPFTLHH38 pKa = 6.49LVEE41 pKa = 4.57FTSGVVSCPTALPATGRR58 pKa = 11.84ARR60 pKa = 11.84DD61 pKa = 3.72DD62 pKa = 3.6DD63 pKa = 4.09DD64 pKa = 4.94FRR66 pKa = 11.84FEE68 pKa = 4.26VTGLLTNQQLQVAQATPIFDD88 pKa = 3.61VYY90 pKa = 9.88LTEE93 pKa = 3.96VSRR96 pKa = 11.84DD97 pKa = 3.47LGHH100 pKa = 6.94CSSGFHH106 pKa = 6.87RR107 pKa = 11.84GNSLAEE113 pKa = 3.84IAPVSLRR120 pKa = 11.84RR121 pKa = 11.84NAQGKK126 pKa = 8.97
Molecular weight: 13.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9IG43|ORFC4_PICV Uncharacterized protein ORFC4 OS=Pigeon circovirus OX=126070 GN=ORFC4 PE=4 SV=1
MM1 pKa = 7.41RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84FRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84APIRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84IRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84TRR24 pKa = 11.84LSRR27 pKa = 11.84NIRR30 pKa = 11.84GHH32 pKa = 5.74RR33 pKa = 11.84RR34 pKa = 11.84SSRR37 pKa = 11.84IYY39 pKa = 9.05YY40 pKa = 9.71FRR42 pKa = 11.84LRR44 pKa = 11.84RR45 pKa = 11.84KK46 pKa = 10.34DD47 pKa = 4.22KK48 pKa = 10.09ITLTQATNDD57 pKa = 3.58FKK59 pKa = 11.19FGTGIFTFKK68 pKa = 10.66LADD71 pKa = 3.57VLTVGLNAPTLKK83 pKa = 10.78VPFEE87 pKa = 5.07DD88 pKa = 3.91YY89 pKa = 10.86QIALVKK95 pKa = 10.82VEE97 pKa = 4.09MRR99 pKa = 11.84PLGVDD104 pKa = 2.6ITTWKK109 pKa = 10.66GFGHH113 pKa = 5.88TVPMYY118 pKa = 10.29DD119 pKa = 3.58ARR121 pKa = 11.84LKK123 pKa = 9.52TFQGQVDD130 pKa = 4.39LGDD133 pKa = 4.67DD134 pKa = 3.71PLMDD138 pKa = 4.22FDD140 pKa = 6.26GARR143 pKa = 11.84KK144 pKa = 8.46WDD146 pKa = 3.5LRR148 pKa = 11.84KK149 pKa = 10.26GFKK152 pKa = 10.14RR153 pKa = 11.84LIRR156 pKa = 11.84PRR158 pKa = 11.84PQLTIADD165 pKa = 4.52LATANQSAATWFSGRR180 pKa = 11.84NQWIPLQVSGNSLFPQKK197 pKa = 10.63VNHH200 pKa = 6.35YY201 pKa = 9.13GLAFSYY207 pKa = 9.02LQPQPDD213 pKa = 3.39PMYY216 pKa = 10.0YY217 pKa = 8.59EE218 pKa = 4.78CEE220 pKa = 3.75VTFYY224 pKa = 11.71VKK226 pKa = 10.49FRR228 pKa = 11.84QFAWTTLNVPPTPNIEE244 pKa = 4.11GMEE247 pKa = 4.2LMHH250 pKa = 7.05ICNGDD255 pKa = 3.82CNQCFADD262 pKa = 5.3ALDD265 pKa = 4.73PDD267 pKa = 4.17SAVDD271 pKa = 3.97SEE273 pKa = 4.46
MM1 pKa = 7.41RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84FRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84APIRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84IRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84TRR24 pKa = 11.84LSRR27 pKa = 11.84NIRR30 pKa = 11.84GHH32 pKa = 5.74RR33 pKa = 11.84RR34 pKa = 11.84SSRR37 pKa = 11.84IYY39 pKa = 9.05YY40 pKa = 9.71FRR42 pKa = 11.84LRR44 pKa = 11.84RR45 pKa = 11.84KK46 pKa = 10.34DD47 pKa = 4.22KK48 pKa = 10.09ITLTQATNDD57 pKa = 3.58FKK59 pKa = 11.19FGTGIFTFKK68 pKa = 10.66LADD71 pKa = 3.57VLTVGLNAPTLKK83 pKa = 10.78VPFEE87 pKa = 5.07DD88 pKa = 3.91YY89 pKa = 10.86QIALVKK95 pKa = 10.82VEE97 pKa = 4.09MRR99 pKa = 11.84PLGVDD104 pKa = 2.6ITTWKK109 pKa = 10.66GFGHH113 pKa = 5.88TVPMYY118 pKa = 10.29DD119 pKa = 3.58ARR121 pKa = 11.84LKK123 pKa = 9.52TFQGQVDD130 pKa = 4.39LGDD133 pKa = 4.67DD134 pKa = 3.71PLMDD138 pKa = 4.22FDD140 pKa = 6.26GARR143 pKa = 11.84KK144 pKa = 8.46WDD146 pKa = 3.5LRR148 pKa = 11.84KK149 pKa = 10.26GFKK152 pKa = 10.14RR153 pKa = 11.84LIRR156 pKa = 11.84PRR158 pKa = 11.84PQLTIADD165 pKa = 4.52LATANQSAATWFSGRR180 pKa = 11.84NQWIPLQVSGNSLFPQKK197 pKa = 10.63VNHH200 pKa = 6.35YY201 pKa = 9.13GLAFSYY207 pKa = 9.02LQPQPDD213 pKa = 3.39PMYY216 pKa = 10.0YY217 pKa = 8.59EE218 pKa = 4.78CEE220 pKa = 3.75VTFYY224 pKa = 11.71VKK226 pKa = 10.49FRR228 pKa = 11.84QFAWTTLNVPPTPNIEE244 pKa = 4.11GMEE247 pKa = 4.2LMHH250 pKa = 7.05ICNGDD255 pKa = 3.82CNQCFADD262 pKa = 5.3ALDD265 pKa = 4.73PDD267 pKa = 4.17SAVDD271 pKa = 3.97SEE273 pKa = 4.46
Molecular weight: 31.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
895 |
82 |
317 |
179.0 |
20.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.598 ± 1.026 | 2.011 ± 0.325 |
5.14 ± 1.02 | 4.358 ± 1.328 |
5.363 ± 0.551 | 7.821 ± 1.031 |
2.458 ± 0.486 | 3.911 ± 0.453 |
5.14 ± 1.642 | 8.38 ± 0.825 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.676 ± 0.335 | 3.24 ± 0.328 |
6.145 ± 0.611 | 4.469 ± 0.876 |
8.492 ± 1.448 | 6.145 ± 2.26 |
5.922 ± 1.165 | 6.816 ± 1.098 |
2.011 ± 0.561 | 2.905 ± 0.536 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
