
gamma proteobacterium BDW918
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Spongiibacteraceae; unclassified Spongiibacteraceae
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
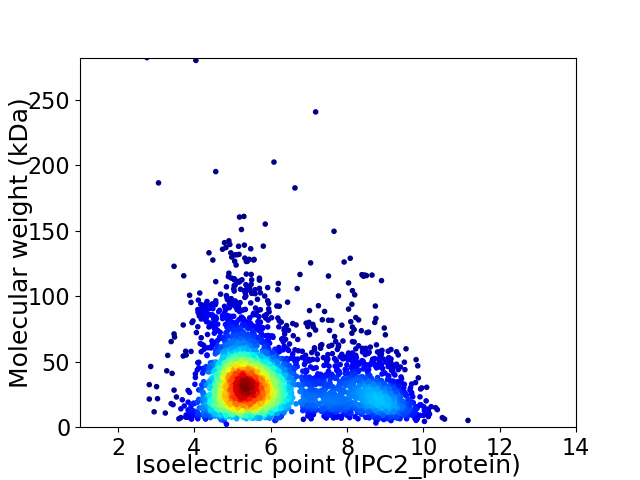
Virtual 2D-PAGE plot for 3794 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I2JF95|I2JF95_9GAMM Uncharacterized protein OS=gamma proteobacterium BDW918 OX=1168065 GN=DOK_17700 PE=4 SV=1
MM1 pKa = 7.35FMSKK5 pKa = 7.11QWKK8 pKa = 8.35SALAAGVILASATGAAMAEE27 pKa = 4.07DD28 pKa = 4.45QEE30 pKa = 4.7TGFYY34 pKa = 10.67VGGNAFYY41 pKa = 11.4NEE43 pKa = 4.05VFDD46 pKa = 5.58ADD48 pKa = 3.78GTATTTTAGGFAAIPLIGDD67 pKa = 3.68VLNGLLGGSASSSANLEE84 pKa = 3.83TSYY87 pKa = 11.71DD88 pKa = 3.41NDD90 pKa = 2.76ISYY93 pKa = 10.75GFSVGYY99 pKa = 10.28KK100 pKa = 8.41FASPVRR106 pKa = 11.84IEE108 pKa = 3.61FEE110 pKa = 4.07YY111 pKa = 10.88RR112 pKa = 11.84QGEE115 pKa = 4.1NDD117 pKa = 2.9VDD119 pKa = 4.49KK120 pKa = 10.76ISSGGVSTTSDD131 pKa = 2.79SSLEE135 pKa = 4.16VTSMMGNVWYY145 pKa = 10.07DD146 pKa = 3.87FAAGEE151 pKa = 4.15RR152 pKa = 11.84LRR154 pKa = 11.84PYY156 pKa = 10.26IGFGLGMANLDD167 pKa = 4.06MGDD170 pKa = 3.87ADD172 pKa = 4.91DD173 pKa = 5.97DD174 pKa = 4.19VTIGQLGAGITYY186 pKa = 10.09FLTPRR191 pKa = 11.84LALDD195 pKa = 3.55AGYY198 pKa = 9.74RR199 pKa = 11.84YY200 pKa = 10.16SVAEE204 pKa = 4.05DD205 pKa = 3.42ATFKK209 pKa = 10.93SATIEE214 pKa = 3.97QEE216 pKa = 3.96NEE218 pKa = 3.46YY219 pKa = 10.38SAQSVQVGLRR229 pKa = 11.84YY230 pKa = 10.39NFFEE234 pKa = 4.1AQYY237 pKa = 10.75GVKK240 pKa = 10.47DD241 pKa = 3.55SDD243 pKa = 3.82GDD245 pKa = 4.18GVSDD249 pKa = 4.51EE250 pKa = 4.4TDD252 pKa = 3.23EE253 pKa = 5.44CPGTPRR259 pKa = 11.84GVQVDD264 pKa = 4.07SVGCPLDD271 pKa = 3.81GDD273 pKa = 3.96NDD275 pKa = 3.97GVADD279 pKa = 5.15YY280 pKa = 11.39LDD282 pKa = 3.62QCPNTPAGAKK292 pKa = 9.73VNASGCPLDD301 pKa = 4.24GDD303 pKa = 3.96NDD305 pKa = 4.07GVADD309 pKa = 5.33ADD311 pKa = 4.43DD312 pKa = 4.68ACPDD316 pKa = 3.72TPAGQAVMSNGCAKK330 pKa = 10.3DD331 pKa = 3.15QAVILRR337 pKa = 11.84GVNFEE342 pKa = 4.81LNSAQLTMNAEE353 pKa = 4.31TILNGVAEE361 pKa = 4.68TLVSSPGFDD370 pKa = 3.81VEE372 pKa = 4.64LQGHH376 pKa = 5.35TDD378 pKa = 3.22STGSDD383 pKa = 3.75SYY385 pKa = 12.21NMNLSQNRR393 pKa = 11.84AKK395 pKa = 10.43SVKK398 pKa = 10.02SYY400 pKa = 10.5LVSSGVEE407 pKa = 4.05ANRR410 pKa = 11.84LTARR414 pKa = 11.84GYY416 pKa = 11.14GEE418 pKa = 4.18EE419 pKa = 4.23QPIASNDD426 pKa = 3.75TKK428 pKa = 10.96DD429 pKa = 3.43GRR431 pKa = 11.84AQNRR435 pKa = 11.84RR436 pKa = 11.84VEE438 pKa = 4.27LKK440 pKa = 10.88VIGDD444 pKa = 4.09GSSDD448 pKa = 3.43SVEE451 pKa = 3.37PMMYY455 pKa = 10.04DD456 pKa = 2.98EE457 pKa = 5.38PMVEE461 pKa = 4.24EE462 pKa = 4.38PAVEE466 pKa = 4.33EE467 pKa = 4.14PMMDD471 pKa = 3.05EE472 pKa = 4.2PMVEE476 pKa = 4.22EE477 pKa = 4.26EE478 pKa = 4.88PMVEE482 pKa = 4.31DD483 pKa = 3.84EE484 pKa = 5.0PMMEE488 pKa = 4.13EE489 pKa = 4.28APAEE493 pKa = 4.2AEE495 pKa = 3.79PSVEE499 pKa = 5.52DD500 pKa = 4.11EE501 pKa = 4.39YY502 pKa = 11.28EE503 pKa = 4.02YY504 pKa = 11.37DD505 pKa = 3.64PSAEE509 pKa = 3.57TDD511 pKa = 3.68YY512 pKa = 11.98
MM1 pKa = 7.35FMSKK5 pKa = 7.11QWKK8 pKa = 8.35SALAAGVILASATGAAMAEE27 pKa = 4.07DD28 pKa = 4.45QEE30 pKa = 4.7TGFYY34 pKa = 10.67VGGNAFYY41 pKa = 11.4NEE43 pKa = 4.05VFDD46 pKa = 5.58ADD48 pKa = 3.78GTATTTTAGGFAAIPLIGDD67 pKa = 3.68VLNGLLGGSASSSANLEE84 pKa = 3.83TSYY87 pKa = 11.71DD88 pKa = 3.41NDD90 pKa = 2.76ISYY93 pKa = 10.75GFSVGYY99 pKa = 10.28KK100 pKa = 8.41FASPVRR106 pKa = 11.84IEE108 pKa = 3.61FEE110 pKa = 4.07YY111 pKa = 10.88RR112 pKa = 11.84QGEE115 pKa = 4.1NDD117 pKa = 2.9VDD119 pKa = 4.49KK120 pKa = 10.76ISSGGVSTTSDD131 pKa = 2.79SSLEE135 pKa = 4.16VTSMMGNVWYY145 pKa = 10.07DD146 pKa = 3.87FAAGEE151 pKa = 4.15RR152 pKa = 11.84LRR154 pKa = 11.84PYY156 pKa = 10.26IGFGLGMANLDD167 pKa = 4.06MGDD170 pKa = 3.87ADD172 pKa = 4.91DD173 pKa = 5.97DD174 pKa = 4.19VTIGQLGAGITYY186 pKa = 10.09FLTPRR191 pKa = 11.84LALDD195 pKa = 3.55AGYY198 pKa = 9.74RR199 pKa = 11.84YY200 pKa = 10.16SVAEE204 pKa = 4.05DD205 pKa = 3.42ATFKK209 pKa = 10.93SATIEE214 pKa = 3.97QEE216 pKa = 3.96NEE218 pKa = 3.46YY219 pKa = 10.38SAQSVQVGLRR229 pKa = 11.84YY230 pKa = 10.39NFFEE234 pKa = 4.1AQYY237 pKa = 10.75GVKK240 pKa = 10.47DD241 pKa = 3.55SDD243 pKa = 3.82GDD245 pKa = 4.18GVSDD249 pKa = 4.51EE250 pKa = 4.4TDD252 pKa = 3.23EE253 pKa = 5.44CPGTPRR259 pKa = 11.84GVQVDD264 pKa = 4.07SVGCPLDD271 pKa = 3.81GDD273 pKa = 3.96NDD275 pKa = 3.97GVADD279 pKa = 5.15YY280 pKa = 11.39LDD282 pKa = 3.62QCPNTPAGAKK292 pKa = 9.73VNASGCPLDD301 pKa = 4.24GDD303 pKa = 3.96NDD305 pKa = 4.07GVADD309 pKa = 5.33ADD311 pKa = 4.43DD312 pKa = 4.68ACPDD316 pKa = 3.72TPAGQAVMSNGCAKK330 pKa = 10.3DD331 pKa = 3.15QAVILRR337 pKa = 11.84GVNFEE342 pKa = 4.81LNSAQLTMNAEE353 pKa = 4.31TILNGVAEE361 pKa = 4.68TLVSSPGFDD370 pKa = 3.81VEE372 pKa = 4.64LQGHH376 pKa = 5.35TDD378 pKa = 3.22STGSDD383 pKa = 3.75SYY385 pKa = 12.21NMNLSQNRR393 pKa = 11.84AKK395 pKa = 10.43SVKK398 pKa = 10.02SYY400 pKa = 10.5LVSSGVEE407 pKa = 4.05ANRR410 pKa = 11.84LTARR414 pKa = 11.84GYY416 pKa = 11.14GEE418 pKa = 4.18EE419 pKa = 4.23QPIASNDD426 pKa = 3.75TKK428 pKa = 10.96DD429 pKa = 3.43GRR431 pKa = 11.84AQNRR435 pKa = 11.84RR436 pKa = 11.84VEE438 pKa = 4.27LKK440 pKa = 10.88VIGDD444 pKa = 4.09GSSDD448 pKa = 3.43SVEE451 pKa = 3.37PMMYY455 pKa = 10.04DD456 pKa = 2.98EE457 pKa = 5.38PMVEE461 pKa = 4.24EE462 pKa = 4.38PAVEE466 pKa = 4.33EE467 pKa = 4.14PMMDD471 pKa = 3.05EE472 pKa = 4.2PMVEE476 pKa = 4.22EE477 pKa = 4.26EE478 pKa = 4.88PMVEE482 pKa = 4.31DD483 pKa = 3.84EE484 pKa = 5.0PMMEE488 pKa = 4.13EE489 pKa = 4.28APAEE493 pKa = 4.2AEE495 pKa = 3.79PSVEE499 pKa = 5.52DD500 pKa = 4.11EE501 pKa = 4.39YY502 pKa = 11.28EE503 pKa = 4.02YY504 pKa = 11.37DD505 pKa = 3.64PSAEE509 pKa = 3.57TDD511 pKa = 3.68YY512 pKa = 11.98
Molecular weight: 54.33 kDa
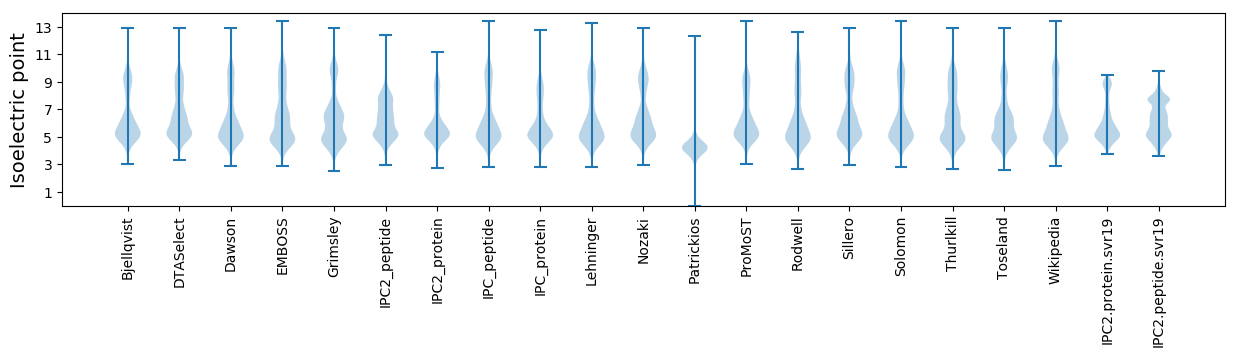
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I2JER4|I2JER4_9GAMM Putative cytochrome oxidase assembly protein OS=gamma proteobacterium BDW918 OX=1168065 GN=DOK_18340 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.0KK13 pKa = 10.58ARR15 pKa = 11.84NHH17 pKa = 5.3GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.45NGRR29 pKa = 11.84KK30 pKa = 9.38LINRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.31GRR40 pKa = 11.84AVLSAA45 pKa = 3.96
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.0KK13 pKa = 10.58ARR15 pKa = 11.84NHH17 pKa = 5.3GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.45NGRR29 pKa = 11.84KK30 pKa = 9.38LINRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.31GRR40 pKa = 11.84AVLSAA45 pKa = 3.96
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1259801 |
20 |
2945 |
332.1 |
36.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.344 ± 0.046 | 1.106 ± 0.014 |
5.813 ± 0.034 | 5.874 ± 0.031 |
3.838 ± 0.023 | 7.522 ± 0.038 |
2.158 ± 0.019 | 5.846 ± 0.027 |
4.215 ± 0.036 | 10.537 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.023 | 3.8 ± 0.029 |
4.292 ± 0.026 | 4.095 ± 0.027 |
5.585 ± 0.038 | 6.678 ± 0.037 |
4.898 ± 0.043 | 6.809 ± 0.034 |
1.26 ± 0.016 | 2.899 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
