
Bos taurus papillomavirus 18
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyokappapapillomavirus; Dyokappapapillomavirus 4
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
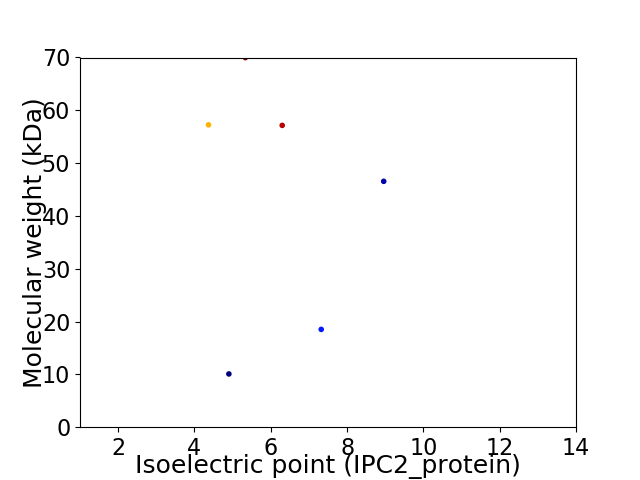
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B2K2B2|A0A1B2K2B2_9PAPI Minor capsid protein L2 OS=Bos taurus papillomavirus 18 OX=1887216 GN=L2 PE=3 SV=1
MM1 pKa = 7.33ATNKK5 pKa = 9.88RR6 pKa = 11.84SRR8 pKa = 11.84VKK10 pKa = 10.24RR11 pKa = 11.84ASVSDD16 pKa = 3.82LAKK19 pKa = 9.71TCATGDD25 pKa = 3.91CPEE28 pKa = 4.23DD29 pKa = 3.57VYY31 pKa = 12.03NKK33 pKa = 8.78VTGNTLADD41 pKa = 4.14KK42 pKa = 10.41ILRR45 pKa = 11.84FLSPFLYY52 pKa = 10.55FGGLSIGTGAGGGGRR67 pKa = 11.84FGYY70 pKa = 10.22RR71 pKa = 11.84PIGGTSGGGRR81 pKa = 11.84PTVTVRR87 pKa = 11.84PVVPVEE93 pKa = 4.29TIAPEE98 pKa = 3.93IEE100 pKa = 4.15VGGTIDD106 pKa = 3.28SSGVIDD112 pKa = 3.66AAGPSVIEE120 pKa = 3.85LTPINGDD127 pKa = 3.28PTVIDD132 pKa = 3.91VAPDD136 pKa = 3.27TAINEE141 pKa = 4.27TGPPILEE148 pKa = 4.04VTPEE152 pKa = 3.74VRR154 pKa = 11.84VTVSGRR160 pKa = 11.84EE161 pKa = 4.07TEE163 pKa = 4.41TVVQNPTFTPDD174 pKa = 3.07PSTINTSHH182 pKa = 6.7NEE184 pKa = 4.09NIVFEE189 pKa = 4.82PNTNAVYY196 pKa = 10.53VGSTAEE202 pKa = 3.83TSEE205 pKa = 4.36EE206 pKa = 3.86IEE208 pKa = 3.97LSVFSRR214 pKa = 11.84GDD216 pKa = 3.31TFSDD220 pKa = 3.49SFATEE225 pKa = 3.96VEE227 pKa = 4.38EE228 pKa = 4.34TSFDD232 pKa = 3.29ASTPLVRR239 pKa = 11.84QAEE242 pKa = 4.21GRR244 pKa = 11.84PINRR248 pKa = 11.84FGRR251 pKa = 11.84YY252 pKa = 6.77FQQVPVSEE260 pKa = 4.82PEE262 pKa = 4.06FLSRR266 pKa = 11.84PASMVTFGNAAYY278 pKa = 10.33DD279 pKa = 3.88PDD281 pKa = 3.46ATAIFEE287 pKa = 4.8DD288 pKa = 4.48DD289 pKa = 4.45LRR291 pKa = 11.84DD292 pKa = 4.03LEE294 pKa = 5.57DD295 pKa = 3.55IAAAPYY301 pKa = 10.54TDD303 pKa = 5.01FSDD306 pKa = 4.37LKK308 pKa = 9.09TLSRR312 pKa = 11.84PYY314 pKa = 9.78YY315 pKa = 9.92SRR317 pKa = 11.84GGRR320 pKa = 11.84TGRR323 pKa = 11.84LIVSRR328 pKa = 11.84VGTKK332 pKa = 8.23STIHH336 pKa = 5.6TRR338 pKa = 11.84SGLSIGASVHH348 pKa = 6.23YY349 pKa = 8.72YY350 pKa = 10.24TEE352 pKa = 4.49LSTINEE358 pKa = 4.21PTVAEE363 pKa = 4.04QPPFEE368 pKa = 5.46ISDD371 pKa = 3.95FVGSTSADD379 pKa = 3.16EE380 pKa = 5.07AFINSQGTEE389 pKa = 3.95EE390 pKa = 4.76FEE392 pKa = 5.07LISLRR397 pKa = 11.84DD398 pKa = 3.71PSVEE402 pKa = 4.09TLDD405 pKa = 3.55EE406 pKa = 4.06TSEE409 pKa = 4.11YY410 pKa = 11.03SEE412 pKa = 4.32TALLDD417 pKa = 4.25HH418 pKa = 7.22IEE420 pKa = 4.52EE421 pKa = 4.19IGEE424 pKa = 3.99NLRR427 pKa = 11.84LIIQSDD433 pKa = 3.54EE434 pKa = 4.23RR435 pKa = 11.84EE436 pKa = 3.84QTIISVPSFNEE447 pKa = 3.98SVAEE451 pKa = 3.93NAVSIVVDD459 pKa = 3.81YY460 pKa = 10.77PNSTSEE466 pKa = 4.38SPATNYY472 pKa = 9.75PMEE475 pKa = 4.63YY476 pKa = 8.14PTDD479 pKa = 4.16TISPSQPATPGLTPLIPSSLQPTNYY504 pKa = 9.5FDD506 pKa = 3.22YY507 pKa = 9.85WQLFEE512 pKa = 4.52PSLWRR517 pKa = 11.84SRR519 pKa = 11.84KK520 pKa = 8.93RR521 pKa = 11.84KK522 pKa = 9.47RR523 pKa = 11.84NVYY526 pKa = 9.2YY527 pKa = 10.94
MM1 pKa = 7.33ATNKK5 pKa = 9.88RR6 pKa = 11.84SRR8 pKa = 11.84VKK10 pKa = 10.24RR11 pKa = 11.84ASVSDD16 pKa = 3.82LAKK19 pKa = 9.71TCATGDD25 pKa = 3.91CPEE28 pKa = 4.23DD29 pKa = 3.57VYY31 pKa = 12.03NKK33 pKa = 8.78VTGNTLADD41 pKa = 4.14KK42 pKa = 10.41ILRR45 pKa = 11.84FLSPFLYY52 pKa = 10.55FGGLSIGTGAGGGGRR67 pKa = 11.84FGYY70 pKa = 10.22RR71 pKa = 11.84PIGGTSGGGRR81 pKa = 11.84PTVTVRR87 pKa = 11.84PVVPVEE93 pKa = 4.29TIAPEE98 pKa = 3.93IEE100 pKa = 4.15VGGTIDD106 pKa = 3.28SSGVIDD112 pKa = 3.66AAGPSVIEE120 pKa = 3.85LTPINGDD127 pKa = 3.28PTVIDD132 pKa = 3.91VAPDD136 pKa = 3.27TAINEE141 pKa = 4.27TGPPILEE148 pKa = 4.04VTPEE152 pKa = 3.74VRR154 pKa = 11.84VTVSGRR160 pKa = 11.84EE161 pKa = 4.07TEE163 pKa = 4.41TVVQNPTFTPDD174 pKa = 3.07PSTINTSHH182 pKa = 6.7NEE184 pKa = 4.09NIVFEE189 pKa = 4.82PNTNAVYY196 pKa = 10.53VGSTAEE202 pKa = 3.83TSEE205 pKa = 4.36EE206 pKa = 3.86IEE208 pKa = 3.97LSVFSRR214 pKa = 11.84GDD216 pKa = 3.31TFSDD220 pKa = 3.49SFATEE225 pKa = 3.96VEE227 pKa = 4.38EE228 pKa = 4.34TSFDD232 pKa = 3.29ASTPLVRR239 pKa = 11.84QAEE242 pKa = 4.21GRR244 pKa = 11.84PINRR248 pKa = 11.84FGRR251 pKa = 11.84YY252 pKa = 6.77FQQVPVSEE260 pKa = 4.82PEE262 pKa = 4.06FLSRR266 pKa = 11.84PASMVTFGNAAYY278 pKa = 10.33DD279 pKa = 3.88PDD281 pKa = 3.46ATAIFEE287 pKa = 4.8DD288 pKa = 4.48DD289 pKa = 4.45LRR291 pKa = 11.84DD292 pKa = 4.03LEE294 pKa = 5.57DD295 pKa = 3.55IAAAPYY301 pKa = 10.54TDD303 pKa = 5.01FSDD306 pKa = 4.37LKK308 pKa = 9.09TLSRR312 pKa = 11.84PYY314 pKa = 9.78YY315 pKa = 9.92SRR317 pKa = 11.84GGRR320 pKa = 11.84TGRR323 pKa = 11.84LIVSRR328 pKa = 11.84VGTKK332 pKa = 8.23STIHH336 pKa = 5.6TRR338 pKa = 11.84SGLSIGASVHH348 pKa = 6.23YY349 pKa = 8.72YY350 pKa = 10.24TEE352 pKa = 4.49LSTINEE358 pKa = 4.21PTVAEE363 pKa = 4.04QPPFEE368 pKa = 5.46ISDD371 pKa = 3.95FVGSTSADD379 pKa = 3.16EE380 pKa = 5.07AFINSQGTEE389 pKa = 3.95EE390 pKa = 4.76FEE392 pKa = 5.07LISLRR397 pKa = 11.84DD398 pKa = 3.71PSVEE402 pKa = 4.09TLDD405 pKa = 3.55EE406 pKa = 4.06TSEE409 pKa = 4.11YY410 pKa = 11.03SEE412 pKa = 4.32TALLDD417 pKa = 4.25HH418 pKa = 7.22IEE420 pKa = 4.52EE421 pKa = 4.19IGEE424 pKa = 3.99NLRR427 pKa = 11.84LIIQSDD433 pKa = 3.54EE434 pKa = 4.23RR435 pKa = 11.84EE436 pKa = 3.84QTIISVPSFNEE447 pKa = 3.98SVAEE451 pKa = 3.93NAVSIVVDD459 pKa = 3.81YY460 pKa = 10.77PNSTSEE466 pKa = 4.38SPATNYY472 pKa = 9.75PMEE475 pKa = 4.63YY476 pKa = 8.14PTDD479 pKa = 4.16TISPSQPATPGLTPLIPSSLQPTNYY504 pKa = 9.5FDD506 pKa = 3.22YY507 pKa = 9.85WQLFEE512 pKa = 4.52PSLWRR517 pKa = 11.84SRR519 pKa = 11.84KK520 pKa = 8.93RR521 pKa = 11.84KK522 pKa = 9.47RR523 pKa = 11.84NVYY526 pKa = 9.2YY527 pKa = 10.94
Molecular weight: 57.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2K215|A0A1B2K215_9PAPI Protein E6 OS=Bos taurus papillomavirus 18 OX=1887216 GN=E6 PE=3 SV=1
MM1 pKa = 7.07EE2 pKa = 4.86TLLSRR7 pKa = 11.84LDD9 pKa = 3.54AVRR12 pKa = 11.84EE13 pKa = 3.97EE14 pKa = 4.81LLTLYY19 pKa = 10.65EE20 pKa = 4.41SGSNRR25 pKa = 11.84LEE27 pKa = 3.82DD28 pKa = 5.03QIRR31 pKa = 11.84HH32 pKa = 4.56WKK34 pKa = 9.73LLRR37 pKa = 11.84KK38 pKa = 9.34EE39 pKa = 3.87YY40 pKa = 11.36VMLHH44 pKa = 6.37FARR47 pKa = 11.84KK48 pKa = 8.93HH49 pKa = 5.38DD50 pKa = 3.68IYY52 pKa = 11.11RR53 pKa = 11.84LGLSSVPTLQVSQAKK68 pKa = 9.69AKK70 pKa = 9.82EE71 pKa = 4.19AIRR74 pKa = 11.84MEE76 pKa = 4.35LLLEE80 pKa = 4.17SLAKK84 pKa = 9.99SEE86 pKa = 4.36YY87 pKa = 10.58SSEE90 pKa = 3.82PWTLSEE96 pKa = 4.3TSIEE100 pKa = 4.1MLEE103 pKa = 4.44TPPKK107 pKa = 10.8DD108 pKa = 3.29CFKK111 pKa = 10.77KK112 pKa = 10.07HH113 pKa = 4.81GVTVEE118 pKa = 3.83VLFDD122 pKa = 4.11GDD124 pKa = 4.06PEE126 pKa = 4.36NVMTYY131 pKa = 8.78TNWKK135 pKa = 9.42DD136 pKa = 2.76IYY138 pKa = 10.28YY139 pKa = 8.17QTEE142 pKa = 4.27SGWEE146 pKa = 3.91KK147 pKa = 8.31TTGHH151 pKa = 5.74VNYY154 pKa = 9.89DD155 pKa = 3.06GCFYY159 pKa = 11.23YY160 pKa = 10.36EE161 pKa = 4.55GRR163 pKa = 11.84SQVYY167 pKa = 7.43FVRR170 pKa = 11.84FAPDD174 pKa = 3.0AARR177 pKa = 11.84YY178 pKa = 9.14GSTGEE183 pKa = 3.89WEE185 pKa = 3.88VRR187 pKa = 11.84YY188 pKa = 10.33QNDD191 pKa = 2.82VFFPVASVSSTSEE204 pKa = 3.43WCPPSTGSGIQPWGDD219 pKa = 3.52KK220 pKa = 11.09LPVDD224 pKa = 4.09TCLGSAGRR232 pKa = 11.84PAQTARR238 pKa = 11.84RR239 pKa = 11.84SPDD242 pKa = 2.99SSGSSATSTTTSRR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84KK259 pKa = 9.29SVRR262 pKa = 11.84IRR264 pKa = 11.84KK265 pKa = 7.86ATAPQEE271 pKa = 4.31AQRR274 pKa = 11.84SSPRR278 pKa = 11.84SSSGAEE284 pKa = 4.12DD285 pKa = 3.3GRR287 pKa = 11.84QARR290 pKa = 11.84FQPGGRR296 pKa = 11.84IPPGSHH302 pKa = 4.05QQVEE306 pKa = 4.32RR307 pKa = 11.84GARR310 pKa = 11.84EE311 pKa = 3.59NPRR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84SRR319 pKa = 11.84LIQLLADD326 pKa = 3.96AADD329 pKa = 3.87PPIAVLRR336 pKa = 11.84GASNSLKK343 pKa = 10.15CLRR346 pKa = 11.84YY347 pKa = 9.45RR348 pKa = 11.84LNRR351 pKa = 11.84RR352 pKa = 11.84HH353 pKa = 6.5RR354 pKa = 11.84GLFNHH359 pKa = 7.45ISTTWSWADD368 pKa = 3.24EE369 pKa = 4.36RR370 pKa = 11.84GPGANARR377 pKa = 11.84MIIAFTNSTQRR388 pKa = 11.84QTFLDD393 pKa = 4.3TVSLPRR399 pKa = 11.84PITYY403 pKa = 10.31FLGNFSSLL411 pKa = 3.44
MM1 pKa = 7.07EE2 pKa = 4.86TLLSRR7 pKa = 11.84LDD9 pKa = 3.54AVRR12 pKa = 11.84EE13 pKa = 3.97EE14 pKa = 4.81LLTLYY19 pKa = 10.65EE20 pKa = 4.41SGSNRR25 pKa = 11.84LEE27 pKa = 3.82DD28 pKa = 5.03QIRR31 pKa = 11.84HH32 pKa = 4.56WKK34 pKa = 9.73LLRR37 pKa = 11.84KK38 pKa = 9.34EE39 pKa = 3.87YY40 pKa = 11.36VMLHH44 pKa = 6.37FARR47 pKa = 11.84KK48 pKa = 8.93HH49 pKa = 5.38DD50 pKa = 3.68IYY52 pKa = 11.11RR53 pKa = 11.84LGLSSVPTLQVSQAKK68 pKa = 9.69AKK70 pKa = 9.82EE71 pKa = 4.19AIRR74 pKa = 11.84MEE76 pKa = 4.35LLLEE80 pKa = 4.17SLAKK84 pKa = 9.99SEE86 pKa = 4.36YY87 pKa = 10.58SSEE90 pKa = 3.82PWTLSEE96 pKa = 4.3TSIEE100 pKa = 4.1MLEE103 pKa = 4.44TPPKK107 pKa = 10.8DD108 pKa = 3.29CFKK111 pKa = 10.77KK112 pKa = 10.07HH113 pKa = 4.81GVTVEE118 pKa = 3.83VLFDD122 pKa = 4.11GDD124 pKa = 4.06PEE126 pKa = 4.36NVMTYY131 pKa = 8.78TNWKK135 pKa = 9.42DD136 pKa = 2.76IYY138 pKa = 10.28YY139 pKa = 8.17QTEE142 pKa = 4.27SGWEE146 pKa = 3.91KK147 pKa = 8.31TTGHH151 pKa = 5.74VNYY154 pKa = 9.89DD155 pKa = 3.06GCFYY159 pKa = 11.23YY160 pKa = 10.36EE161 pKa = 4.55GRR163 pKa = 11.84SQVYY167 pKa = 7.43FVRR170 pKa = 11.84FAPDD174 pKa = 3.0AARR177 pKa = 11.84YY178 pKa = 9.14GSTGEE183 pKa = 3.89WEE185 pKa = 3.88VRR187 pKa = 11.84YY188 pKa = 10.33QNDD191 pKa = 2.82VFFPVASVSSTSEE204 pKa = 3.43WCPPSTGSGIQPWGDD219 pKa = 3.52KK220 pKa = 11.09LPVDD224 pKa = 4.09TCLGSAGRR232 pKa = 11.84PAQTARR238 pKa = 11.84RR239 pKa = 11.84SPDD242 pKa = 2.99SSGSSATSTTTSRR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84KK259 pKa = 9.29SVRR262 pKa = 11.84IRR264 pKa = 11.84KK265 pKa = 7.86ATAPQEE271 pKa = 4.31AQRR274 pKa = 11.84SSPRR278 pKa = 11.84SSSGAEE284 pKa = 4.12DD285 pKa = 3.3GRR287 pKa = 11.84QARR290 pKa = 11.84FQPGGRR296 pKa = 11.84IPPGSHH302 pKa = 4.05QQVEE306 pKa = 4.32RR307 pKa = 11.84GARR310 pKa = 11.84EE311 pKa = 3.59NPRR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84SRR319 pKa = 11.84LIQLLADD326 pKa = 3.96AADD329 pKa = 3.87PPIAVLRR336 pKa = 11.84GASNSLKK343 pKa = 10.15CLRR346 pKa = 11.84YY347 pKa = 9.45RR348 pKa = 11.84LNRR351 pKa = 11.84RR352 pKa = 11.84HH353 pKa = 6.5RR354 pKa = 11.84GLFNHH359 pKa = 7.45ISTTWSWADD368 pKa = 3.24EE369 pKa = 4.36RR370 pKa = 11.84GPGANARR377 pKa = 11.84MIIAFTNSTQRR388 pKa = 11.84QTFLDD393 pKa = 4.3TVSLPRR399 pKa = 11.84PITYY403 pKa = 10.31FLGNFSSLL411 pKa = 3.44
Molecular weight: 46.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2293 |
88 |
608 |
382.2 |
43.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.582 ± 0.664 | 2.093 ± 0.816 |
5.713 ± 0.368 | 7.021 ± 0.678 |
4.405 ± 0.274 | 5.931 ± 0.73 |
2.268 ± 0.496 | 5.233 ± 0.592 |
4.666 ± 0.936 | 8.112 ± 0.762 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.919 ± 0.458 | 5.015 ± 1.012 |
5.451 ± 0.782 | 4.187 ± 0.638 |
6.106 ± 0.996 | 8.33 ± 0.848 |
6.847 ± 1.056 | 5.975 ± 0.595 |
1.526 ± 0.347 | 3.62 ± 0.249 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
