
Human papillomavirus 107
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 2
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
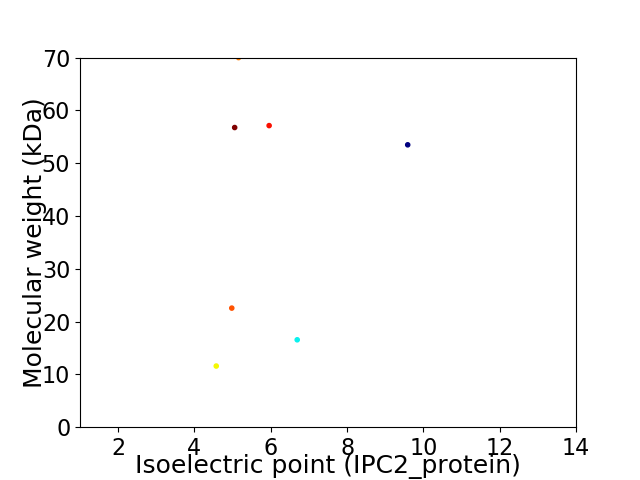
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3RKL8|A3RKL8_9PAPI Replication protein E1 OS=Human papillomavirus 107 OX=427343 GN=E1 PE=3 SV=1
MM1 pKa = 7.43IGKK4 pKa = 8.97EE5 pKa = 3.83VTIPDD10 pKa = 3.41IVLEE14 pKa = 4.05EE15 pKa = 4.22LQEE18 pKa = 3.99LAQPIDD24 pKa = 3.28LHH26 pKa = 8.37CYY28 pKa = 9.94EE29 pKa = 5.38EE30 pKa = 5.35LPEE33 pKa = 5.28LPDD36 pKa = 3.51ATSEE40 pKa = 4.13EE41 pKa = 4.65SEE43 pKa = 4.45EE44 pKa = 4.04EE45 pKa = 4.34PEE47 pKa = 4.39RR48 pKa = 11.84IPYY51 pKa = 9.63KK52 pKa = 10.4IVVPCSGCPVKK63 pKa = 10.75LRR65 pKa = 11.84LYY67 pKa = 10.77VFATHH72 pKa = 6.68FGIRR76 pKa = 11.84ALQDD80 pKa = 3.52LLLQEE85 pKa = 4.41VQLICPQCRR94 pKa = 11.84EE95 pKa = 4.38AIRR98 pKa = 11.84HH99 pKa = 5.56GGQQ102 pKa = 2.81
MM1 pKa = 7.43IGKK4 pKa = 8.97EE5 pKa = 3.83VTIPDD10 pKa = 3.41IVLEE14 pKa = 4.05EE15 pKa = 4.22LQEE18 pKa = 3.99LAQPIDD24 pKa = 3.28LHH26 pKa = 8.37CYY28 pKa = 9.94EE29 pKa = 5.38EE30 pKa = 5.35LPEE33 pKa = 5.28LPDD36 pKa = 3.51ATSEE40 pKa = 4.13EE41 pKa = 4.65SEE43 pKa = 4.45EE44 pKa = 4.04EE45 pKa = 4.34PEE47 pKa = 4.39RR48 pKa = 11.84IPYY51 pKa = 9.63KK52 pKa = 10.4IVVPCSGCPVKK63 pKa = 10.75LRR65 pKa = 11.84LYY67 pKa = 10.77VFATHH72 pKa = 6.68FGIRR76 pKa = 11.84ALQDD80 pKa = 3.52LLLQEE85 pKa = 4.41VQLICPQCRR94 pKa = 11.84EE95 pKa = 4.38AIRR98 pKa = 11.84HH99 pKa = 5.56GGQQ102 pKa = 2.81
Molecular weight: 11.58 kDa
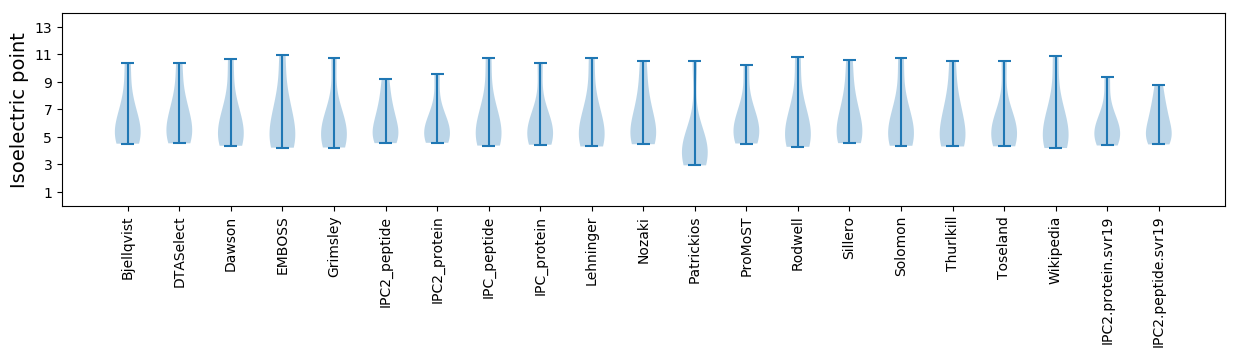
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3RKM0|A3RKM0_9PAPI E4 protein OS=Human papillomavirus 107 OX=427343 GN=E4 PE=4 SV=1
MM1 pKa = 7.02EE2 pKa = 4.6TLSEE6 pKa = 4.42RR7 pKa = 11.84FTALQDD13 pKa = 3.42KK14 pKa = 10.91LMDD17 pKa = 3.74IYY19 pKa = 11.21EE20 pKa = 4.7SGLEE24 pKa = 3.92TLEE27 pKa = 3.88VQIEE31 pKa = 4.05HH32 pKa = 6.09WQLLRR37 pKa = 11.84QEE39 pKa = 4.14QVLLNFARR47 pKa = 11.84RR48 pKa = 11.84NGILRR53 pKa = 11.84LGYY56 pKa = 10.28QPVPSLATSEE66 pKa = 4.44TKK68 pKa = 10.75AKK70 pKa = 10.47DD71 pKa = 3.72AIGMVLMLQSLQNSKK86 pKa = 10.51YY87 pKa = 10.76AEE89 pKa = 4.29EE90 pKa = 4.14TWSLVQTSLEE100 pKa = 4.44TVRR103 pKa = 11.84SPPEE107 pKa = 3.54NCFKK111 pKa = 10.96KK112 pKa = 10.33EE113 pKa = 3.83PQTIEE118 pKa = 3.98VVFDD122 pKa = 3.78GDD124 pKa = 3.59PEE126 pKa = 3.88NRR128 pKa = 11.84MIYY131 pKa = 7.92TVWNWIYY138 pKa = 10.13FQNTEE143 pKa = 5.24DD144 pKa = 2.7IWQKK148 pKa = 10.8VKK150 pKa = 10.8GHH152 pKa = 5.92VDD154 pKa = 3.43YY155 pKa = 11.43EE156 pKa = 4.44GAFYY160 pKa = 10.51MEE162 pKa = 4.83GTHH165 pKa = 4.8KK166 pKa = 10.13HH167 pKa = 5.31YY168 pKa = 10.84YY169 pKa = 9.67IKK171 pKa = 10.76FEE173 pKa = 4.05NDD175 pKa = 2.66ANRR178 pKa = 11.84FSATGLWEE186 pKa = 3.8VHH188 pKa = 5.52VNQDD192 pKa = 3.28TVFTPVTSSTSQAGSRR208 pKa = 11.84LSPLPSDD215 pKa = 3.55PTSRR219 pKa = 11.84RR220 pKa = 11.84PSPSISTVTTKK231 pKa = 9.18QRR233 pKa = 11.84RR234 pKa = 11.84YY235 pKa = 8.53GRR237 pKa = 11.84KK238 pKa = 8.92RR239 pKa = 11.84SSPTATTTGRR249 pKa = 11.84QRR251 pKa = 11.84QRR253 pKa = 11.84TRR255 pKa = 11.84EE256 pKa = 4.05TQQKK260 pKa = 6.95TKK262 pKa = 9.38RR263 pKa = 11.84TRR265 pKa = 11.84SRR267 pKa = 11.84SRR269 pKa = 11.84TKK271 pKa = 10.55SSTRR275 pKa = 11.84SRR277 pKa = 11.84NRR279 pKa = 11.84SRR281 pKa = 11.84SRR283 pKa = 11.84SRR285 pKa = 11.84SSGRR289 pKa = 11.84QDD291 pKa = 2.64TDD293 pKa = 2.9ATKK296 pKa = 10.86GRR298 pKa = 11.84GQGGRR303 pKa = 11.84GRR305 pKa = 11.84EE306 pKa = 3.73RR307 pKa = 11.84EE308 pKa = 3.86RR309 pKa = 11.84EE310 pKa = 3.82RR311 pKa = 11.84EE312 pKa = 4.01RR313 pKa = 11.84EE314 pKa = 3.87RR315 pKa = 11.84EE316 pKa = 3.88RR317 pKa = 11.84ADD319 pKa = 3.21TSNRR323 pKa = 11.84TSRR326 pKa = 11.84GGGRR330 pKa = 11.84SGRR333 pKa = 11.84RR334 pKa = 11.84PQTRR338 pKa = 11.84SRR340 pKa = 11.84SRR342 pKa = 11.84SGGSPDD348 pKa = 3.23RR349 pKa = 11.84VGIAPSEE356 pKa = 4.27VGRR359 pKa = 11.84TLQSVGTTGHH369 pKa = 5.88GRR371 pKa = 11.84LGRR374 pKa = 11.84LLDD377 pKa = 3.81EE378 pKa = 5.48AKK380 pKa = 10.56DD381 pKa = 3.64PPVILLRR388 pKa = 11.84GEE390 pKa = 4.13ANQLKK395 pKa = 9.98CYY397 pKa = 10.28RR398 pKa = 11.84FRR400 pKa = 11.84ARR402 pKa = 11.84KK403 pKa = 8.23KK404 pKa = 9.45HH405 pKa = 5.36KK406 pKa = 10.6GNYY409 pKa = 9.43KK410 pKa = 9.98YY411 pKa = 10.7FSSTWSWIGEE421 pKa = 4.06HH422 pKa = 6.4SNDD425 pKa = 3.59RR426 pKa = 11.84IGRR429 pKa = 11.84ARR431 pKa = 11.84MLVSFTSTHH440 pKa = 4.27QRR442 pKa = 11.84EE443 pKa = 4.28MFLDD447 pKa = 3.92TMKK450 pKa = 10.78LPQGVDD456 pKa = 2.63WSYY459 pKa = 12.19GNFDD463 pKa = 3.58SLL465 pKa = 4.67
MM1 pKa = 7.02EE2 pKa = 4.6TLSEE6 pKa = 4.42RR7 pKa = 11.84FTALQDD13 pKa = 3.42KK14 pKa = 10.91LMDD17 pKa = 3.74IYY19 pKa = 11.21EE20 pKa = 4.7SGLEE24 pKa = 3.92TLEE27 pKa = 3.88VQIEE31 pKa = 4.05HH32 pKa = 6.09WQLLRR37 pKa = 11.84QEE39 pKa = 4.14QVLLNFARR47 pKa = 11.84RR48 pKa = 11.84NGILRR53 pKa = 11.84LGYY56 pKa = 10.28QPVPSLATSEE66 pKa = 4.44TKK68 pKa = 10.75AKK70 pKa = 10.47DD71 pKa = 3.72AIGMVLMLQSLQNSKK86 pKa = 10.51YY87 pKa = 10.76AEE89 pKa = 4.29EE90 pKa = 4.14TWSLVQTSLEE100 pKa = 4.44TVRR103 pKa = 11.84SPPEE107 pKa = 3.54NCFKK111 pKa = 10.96KK112 pKa = 10.33EE113 pKa = 3.83PQTIEE118 pKa = 3.98VVFDD122 pKa = 3.78GDD124 pKa = 3.59PEE126 pKa = 3.88NRR128 pKa = 11.84MIYY131 pKa = 7.92TVWNWIYY138 pKa = 10.13FQNTEE143 pKa = 5.24DD144 pKa = 2.7IWQKK148 pKa = 10.8VKK150 pKa = 10.8GHH152 pKa = 5.92VDD154 pKa = 3.43YY155 pKa = 11.43EE156 pKa = 4.44GAFYY160 pKa = 10.51MEE162 pKa = 4.83GTHH165 pKa = 4.8KK166 pKa = 10.13HH167 pKa = 5.31YY168 pKa = 10.84YY169 pKa = 9.67IKK171 pKa = 10.76FEE173 pKa = 4.05NDD175 pKa = 2.66ANRR178 pKa = 11.84FSATGLWEE186 pKa = 3.8VHH188 pKa = 5.52VNQDD192 pKa = 3.28TVFTPVTSSTSQAGSRR208 pKa = 11.84LSPLPSDD215 pKa = 3.55PTSRR219 pKa = 11.84RR220 pKa = 11.84PSPSISTVTTKK231 pKa = 9.18QRR233 pKa = 11.84RR234 pKa = 11.84YY235 pKa = 8.53GRR237 pKa = 11.84KK238 pKa = 8.92RR239 pKa = 11.84SSPTATTTGRR249 pKa = 11.84QRR251 pKa = 11.84QRR253 pKa = 11.84TRR255 pKa = 11.84EE256 pKa = 4.05TQQKK260 pKa = 6.95TKK262 pKa = 9.38RR263 pKa = 11.84TRR265 pKa = 11.84SRR267 pKa = 11.84SRR269 pKa = 11.84TKK271 pKa = 10.55SSTRR275 pKa = 11.84SRR277 pKa = 11.84NRR279 pKa = 11.84SRR281 pKa = 11.84SRR283 pKa = 11.84SRR285 pKa = 11.84SSGRR289 pKa = 11.84QDD291 pKa = 2.64TDD293 pKa = 2.9ATKK296 pKa = 10.86GRR298 pKa = 11.84GQGGRR303 pKa = 11.84GRR305 pKa = 11.84EE306 pKa = 3.73RR307 pKa = 11.84EE308 pKa = 3.86RR309 pKa = 11.84EE310 pKa = 3.82RR311 pKa = 11.84EE312 pKa = 4.01RR313 pKa = 11.84EE314 pKa = 3.87RR315 pKa = 11.84EE316 pKa = 3.88RR317 pKa = 11.84ADD319 pKa = 3.21TSNRR323 pKa = 11.84TSRR326 pKa = 11.84GGGRR330 pKa = 11.84SGRR333 pKa = 11.84RR334 pKa = 11.84PQTRR338 pKa = 11.84SRR340 pKa = 11.84SRR342 pKa = 11.84SGGSPDD348 pKa = 3.23RR349 pKa = 11.84VGIAPSEE356 pKa = 4.27VGRR359 pKa = 11.84TLQSVGTTGHH369 pKa = 5.88GRR371 pKa = 11.84LGRR374 pKa = 11.84LLDD377 pKa = 3.81EE378 pKa = 5.48AKK380 pKa = 10.56DD381 pKa = 3.64PPVILLRR388 pKa = 11.84GEE390 pKa = 4.13ANQLKK395 pKa = 9.98CYY397 pKa = 10.28RR398 pKa = 11.84FRR400 pKa = 11.84ARR402 pKa = 11.84KK403 pKa = 8.23KK404 pKa = 9.45HH405 pKa = 5.36KK406 pKa = 10.6GNYY409 pKa = 9.43KK410 pKa = 9.98YY411 pKa = 10.7FSSTWSWIGEE421 pKa = 4.06HH422 pKa = 6.4SNDD425 pKa = 3.59RR426 pKa = 11.84IGRR429 pKa = 11.84ARR431 pKa = 11.84MLVSFTSTHH440 pKa = 4.27QRR442 pKa = 11.84EE443 pKa = 4.28MFLDD447 pKa = 3.92TMKK450 pKa = 10.78LPQGVDD456 pKa = 2.63WSYY459 pKa = 12.19GNFDD463 pKa = 3.58SLL465 pKa = 4.67
Molecular weight: 53.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2550 |
102 |
607 |
364.3 |
41.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.451 ± 0.433 | 1.804 ± 0.652 |
5.725 ± 0.527 | 7.176 ± 0.915 |
4.353 ± 0.73 | 6.627 ± 0.871 |
2.0 ± 0.214 | 4.706 ± 0.895 |
4.902 ± 0.76 | 8.667 ± 0.914 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.843 ± 0.527 | 4.275 ± 0.842 |
6.196 ± 1.239 | 5.373 ± 0.275 |
6.863 ± 1.25 | 7.49 ± 0.931 |
6.431 ± 0.812 | 5.882 ± 0.906 |
1.255 ± 0.369 | 2.98 ± 0.271 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
