
Sphingomonas sp. KC8
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
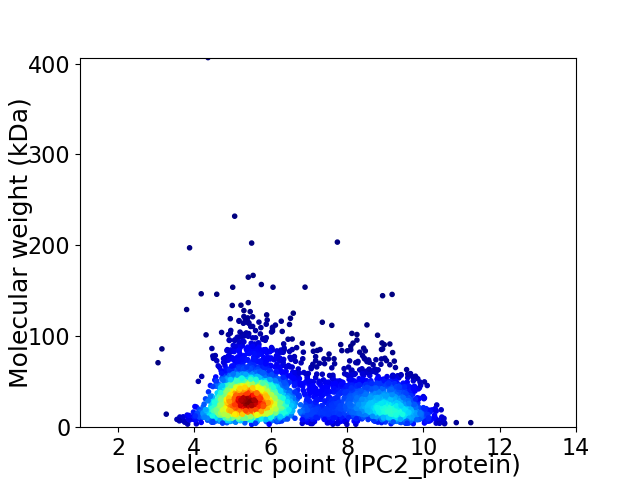
Virtual 2D-PAGE plot for 3846 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9YFJ7|A0A1X9YFJ7_9SPHN Uncharacterized protein OS=Sphingomonas sp. KC8 OX=1030157 GN=KC8_09580 PE=4 SV=1
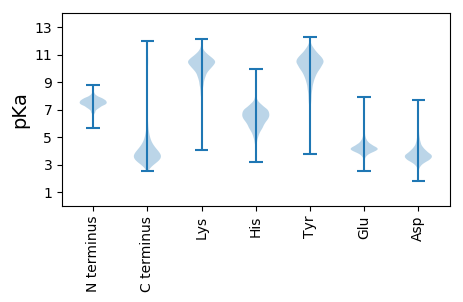
MM1 pKa = 7.28FKK3 pKa = 10.74SSGGGNDD10 pKa = 3.35VLHH13 pKa = 6.88GGDD16 pKa = 3.44GTIVRR21 pKa = 11.84SYY23 pKa = 11.21LSDD26 pKa = 3.38GRR28 pKa = 11.84DD29 pKa = 2.98TVLYY33 pKa = 10.13YY34 pKa = 10.77GYY36 pKa = 10.7DD37 pKa = 3.23SVAIKK42 pKa = 10.64DD43 pKa = 4.51DD44 pKa = 3.99GNHH47 pKa = 5.66WSVEE51 pKa = 3.8KK52 pKa = 10.23TLDD55 pKa = 3.4YY56 pKa = 11.47EE57 pKa = 4.56EE58 pKa = 4.57YY59 pKa = 10.44TDD61 pKa = 3.49TLYY64 pKa = 11.01SIEE67 pKa = 4.32SFGSKK72 pKa = 9.29IFQIINKK79 pKa = 9.04IDD81 pKa = 3.67FSDD84 pKa = 3.74ASSGKK89 pKa = 9.37HH90 pKa = 5.68FKK92 pKa = 10.61DD93 pKa = 3.52ATDD96 pKa = 3.57SDD98 pKa = 3.94IDD100 pKa = 3.84DD101 pKa = 4.08AMEE104 pKa = 4.05TVGDD108 pKa = 4.3GIEE111 pKa = 4.1SFVRR115 pKa = 11.84VDD117 pKa = 3.01VDD119 pKa = 3.99GIEE122 pKa = 4.02VEE124 pKa = 4.38FFNFGTYY131 pKa = 9.72IGTTYY136 pKa = 11.4ADD138 pKa = 3.97RR139 pKa = 11.84FDD141 pKa = 4.54LNQIIGRR148 pKa = 11.84VFDD151 pKa = 4.14GGSGKK156 pKa = 10.19DD157 pKa = 3.25VIDD160 pKa = 3.74YY161 pKa = 11.33SEE163 pKa = 4.43VNDD166 pKa = 3.8TFVVNMRR173 pKa = 11.84PDD175 pKa = 3.05QRR177 pKa = 11.84IAYY180 pKa = 8.55ASSAPSVTDD189 pKa = 3.93SIQSIEE195 pKa = 4.29TVIGAQDD202 pKa = 3.34HH203 pKa = 6.71ANTFVGALGSLDD215 pKa = 4.18EE216 pKa = 4.83NDD218 pKa = 3.44PTFMGGAYY226 pKa = 9.41FANNWGYY233 pKa = 11.57SNLLTYY239 pKa = 10.75VGGSQADD246 pKa = 3.71TYY248 pKa = 11.42EE249 pKa = 4.74FDD251 pKa = 4.27FEE253 pKa = 5.12DD254 pKa = 4.23DD255 pKa = 3.49KK256 pKa = 11.82GVVYY260 pKa = 10.64IRR262 pKa = 11.84DD263 pKa = 3.79NGSGDD268 pKa = 3.56TDD270 pKa = 3.17RR271 pKa = 11.84VAISNVPPGWIDD283 pKa = 3.43EE284 pKa = 4.51STSMHH289 pKa = 6.62HH290 pKa = 6.66GFMVLQNIEE299 pKa = 4.13KK300 pKa = 10.66DD301 pKa = 3.12GDD303 pKa = 4.14NIVSLTIFRR312 pKa = 11.84EE313 pKa = 4.47DD314 pKa = 3.88YY315 pKa = 10.21LTGQSLQLRR324 pKa = 11.84ISSDD328 pKa = 3.03IEE330 pKa = 3.88EE331 pKa = 4.29IEE333 pKa = 4.45INGLTIGMANLLMSMHH349 pKa = 6.36YY350 pKa = 10.6NSGAYY355 pKa = 8.9YY356 pKa = 10.0VAPLDD361 pKa = 4.11AYY363 pKa = 10.99VGLSLARR370 pKa = 11.84LHH372 pKa = 6.18DD373 pKa = 3.74TVYY376 pKa = 11.27NSGPINNGEE385 pKa = 4.26SYY387 pKa = 11.17DD388 pKa = 3.94EE389 pKa = 4.18NLAPTFDD396 pKa = 4.82SNGQSTGVAVSSAIGGTWTTPEE418 pKa = 3.81IATEE422 pKa = 4.2SHH424 pKa = 6.64HH425 pKa = 5.9GWMVTGVTSDD435 pKa = 3.1SGYY438 pKa = 6.97TTLNTTSALKK448 pKa = 10.49EE449 pKa = 3.73IFFAEE454 pKa = 5.65GITAEE459 pKa = 4.39DD460 pKa = 3.51VRR462 pKa = 11.84FTATSSGVDD471 pKa = 3.29NAGLTIHH478 pKa = 7.4IDD480 pKa = 3.21RR481 pKa = 11.84LNYY484 pKa = 9.43SLSVAGFEE492 pKa = 4.64GGRR495 pKa = 11.84TVSGIGFYY503 pKa = 11.02DD504 pKa = 3.74SALHH508 pKa = 5.75AHH510 pKa = 6.32VSSAAAVSLVGTAPGQYY527 pKa = 8.25QVRR530 pKa = 11.84LSGEE534 pKa = 3.84TSYY537 pKa = 11.26TPNAVSATYY546 pKa = 10.09FLEE549 pKa = 4.2ALNFADD555 pKa = 3.88STVIDD560 pKa = 4.98LLNAPLTFKK569 pKa = 10.98GSALNEE575 pKa = 3.97TLYY578 pKa = 11.46GLNTRR583 pKa = 11.84GDD585 pKa = 4.55LIFGLAGNDD594 pKa = 3.52TIYY597 pKa = 10.67GYY599 pKa = 11.25GGADD603 pKa = 3.41VLHH606 pKa = 6.86GGDD609 pKa = 4.41GADD612 pKa = 3.91ALHH615 pKa = 6.81GGDD618 pKa = 5.51GNDD621 pKa = 3.35TLYY624 pKa = 11.45GDD626 pKa = 4.22GGIDD630 pKa = 3.27ALYY633 pKa = 10.42GGEE636 pKa = 4.45GNDD639 pKa = 3.35VLYY642 pKa = 11.19GGDD645 pKa = 4.32GNDD648 pKa = 3.81NLNGEE653 pKa = 4.45NGNDD657 pKa = 3.83TLHH660 pKa = 6.89GGAGNDD666 pKa = 3.47NLRR669 pKa = 11.84GGTGDD674 pKa = 3.44DD675 pKa = 3.46TYY677 pKa = 11.91VFARR681 pKa = 11.84VFSATTAGDD690 pKa = 3.88IIYY693 pKa = 10.34EE694 pKa = 4.07NAADD698 pKa = 4.06GFDD701 pKa = 3.93TIFFTDD707 pKa = 5.12GIEE710 pKa = 4.03ADD712 pKa = 4.46EE713 pKa = 4.63VYY715 pKa = 10.69SWTDD719 pKa = 2.83TSGFLWMQVGGAAATNTLKK738 pKa = 11.07AQGSYY743 pKa = 10.57AAATGITTRR752 pKa = 11.84IEE754 pKa = 3.98QVTFDD759 pKa = 5.32DD760 pKa = 3.79ATVWDD765 pKa = 4.63LTQGLHH771 pKa = 6.52LRR773 pKa = 11.84NNDD776 pKa = 3.29TARR779 pKa = 11.84ALYY782 pKa = 8.02GTIYY786 pKa = 10.89ADD788 pKa = 3.79TIEE791 pKa = 4.92GGSAADD797 pKa = 3.81NIYY800 pKa = 11.1GLGGNDD806 pKa = 3.34TLIGHH811 pKa = 7.42GGNDD815 pKa = 3.69ALRR818 pKa = 11.84GGAGDD823 pKa = 3.68DD824 pKa = 3.54TYY826 pKa = 11.52IFARR830 pKa = 11.84NFSGTTAGDD839 pKa = 3.86IIYY842 pKa = 10.59EE843 pKa = 4.18NVDD846 pKa = 3.5EE847 pKa = 5.03GNDD850 pKa = 3.78TIFFTDD856 pKa = 5.36GIDD859 pKa = 3.44ASEE862 pKa = 4.52VYY864 pKa = 10.25SWTDD868 pKa = 2.81TSGFLWMQVGSAAATNTLKK887 pKa = 11.07AQGSYY892 pKa = 10.87AAVTGITTRR901 pKa = 11.84IEE903 pKa = 3.88QVVFDD908 pKa = 5.87DD909 pKa = 3.73EE910 pKa = 4.98TVWDD914 pKa = 4.24LTQGLHH920 pKa = 6.52LRR922 pKa = 11.84NNDD925 pKa = 3.43TARR928 pKa = 11.84TFYY931 pKa = 9.89GTVYY935 pKa = 11.09GDD937 pKa = 3.59TLEE940 pKa = 4.85GGSASDD946 pKa = 4.27HH947 pKa = 6.57IYY949 pKa = 11.26GLGGNDD955 pKa = 3.34TLIGHH960 pKa = 7.36GGNDD964 pKa = 3.64TLRR967 pKa = 11.84GGDD970 pKa = 3.57GDD972 pKa = 3.91DD973 pKa = 3.5TYY975 pKa = 12.1VFATGFSLNATGDD988 pKa = 4.16RR989 pKa = 11.84IVEE992 pKa = 4.27TATGGTDD999 pKa = 3.43TILFTDD1005 pKa = 5.84GIGADD1010 pKa = 5.57DD1011 pKa = 3.95MFSWTDD1017 pKa = 3.07TSGNLWLQLAANPVSNTLKK1036 pKa = 10.28IDD1038 pKa = 3.65GAYY1041 pKa = 10.01SASTGITTYY1050 pKa = 10.82VEE1052 pKa = 3.95NVVFDD1057 pKa = 5.65DD1058 pKa = 3.53ATAWDD1063 pKa = 4.38LTQGLYY1069 pKa = 10.64LRR1071 pKa = 11.84NNDD1074 pKa = 3.94TGRR1077 pKa = 11.84QMYY1080 pKa = 10.36GSALGDD1086 pKa = 3.86WIEE1089 pKa = 4.57GGAGADD1095 pKa = 3.51TLRR1098 pKa = 11.84GFGGDD1103 pKa = 3.28DD1104 pKa = 3.34TLIGRR1109 pKa = 11.84GGNEE1113 pKa = 3.92DD1114 pKa = 4.11LYY1116 pKa = 11.09GGSGADD1122 pKa = 2.95TFVFRR1127 pKa = 11.84AVDD1130 pKa = 3.4VGNGIDD1136 pKa = 3.59TLRR1139 pKa = 11.84DD1140 pKa = 3.33FSMADD1145 pKa = 3.02NDD1147 pKa = 4.43VIDD1150 pKa = 5.11LRR1152 pKa = 11.84DD1153 pKa = 3.61VLDD1156 pKa = 4.28GGFDD1160 pKa = 3.66PMTDD1164 pKa = 3.18ALSDD1168 pKa = 3.7FVQFTNAGSHH1178 pKa = 4.75SQLRR1182 pKa = 11.84VDD1184 pKa = 4.76LDD1186 pKa = 4.03GAGTLYY1192 pKa = 10.73GWTHH1196 pKa = 6.6IATVNGHH1203 pKa = 5.78TNLDD1207 pKa = 3.64ATTLEE1212 pKa = 4.24ANGYY1216 pKa = 9.43LLAAA1220 pKa = 5.2
MM1 pKa = 7.28FKK3 pKa = 10.74SSGGGNDD10 pKa = 3.35VLHH13 pKa = 6.88GGDD16 pKa = 3.44GTIVRR21 pKa = 11.84SYY23 pKa = 11.21LSDD26 pKa = 3.38GRR28 pKa = 11.84DD29 pKa = 2.98TVLYY33 pKa = 10.13YY34 pKa = 10.77GYY36 pKa = 10.7DD37 pKa = 3.23SVAIKK42 pKa = 10.64DD43 pKa = 4.51DD44 pKa = 3.99GNHH47 pKa = 5.66WSVEE51 pKa = 3.8KK52 pKa = 10.23TLDD55 pKa = 3.4YY56 pKa = 11.47EE57 pKa = 4.56EE58 pKa = 4.57YY59 pKa = 10.44TDD61 pKa = 3.49TLYY64 pKa = 11.01SIEE67 pKa = 4.32SFGSKK72 pKa = 9.29IFQIINKK79 pKa = 9.04IDD81 pKa = 3.67FSDD84 pKa = 3.74ASSGKK89 pKa = 9.37HH90 pKa = 5.68FKK92 pKa = 10.61DD93 pKa = 3.52ATDD96 pKa = 3.57SDD98 pKa = 3.94IDD100 pKa = 3.84DD101 pKa = 4.08AMEE104 pKa = 4.05TVGDD108 pKa = 4.3GIEE111 pKa = 4.1SFVRR115 pKa = 11.84VDD117 pKa = 3.01VDD119 pKa = 3.99GIEE122 pKa = 4.02VEE124 pKa = 4.38FFNFGTYY131 pKa = 9.72IGTTYY136 pKa = 11.4ADD138 pKa = 3.97RR139 pKa = 11.84FDD141 pKa = 4.54LNQIIGRR148 pKa = 11.84VFDD151 pKa = 4.14GGSGKK156 pKa = 10.19DD157 pKa = 3.25VIDD160 pKa = 3.74YY161 pKa = 11.33SEE163 pKa = 4.43VNDD166 pKa = 3.8TFVVNMRR173 pKa = 11.84PDD175 pKa = 3.05QRR177 pKa = 11.84IAYY180 pKa = 8.55ASSAPSVTDD189 pKa = 3.93SIQSIEE195 pKa = 4.29TVIGAQDD202 pKa = 3.34HH203 pKa = 6.71ANTFVGALGSLDD215 pKa = 4.18EE216 pKa = 4.83NDD218 pKa = 3.44PTFMGGAYY226 pKa = 9.41FANNWGYY233 pKa = 11.57SNLLTYY239 pKa = 10.75VGGSQADD246 pKa = 3.71TYY248 pKa = 11.42EE249 pKa = 4.74FDD251 pKa = 4.27FEE253 pKa = 5.12DD254 pKa = 4.23DD255 pKa = 3.49KK256 pKa = 11.82GVVYY260 pKa = 10.64IRR262 pKa = 11.84DD263 pKa = 3.79NGSGDD268 pKa = 3.56TDD270 pKa = 3.17RR271 pKa = 11.84VAISNVPPGWIDD283 pKa = 3.43EE284 pKa = 4.51STSMHH289 pKa = 6.62HH290 pKa = 6.66GFMVLQNIEE299 pKa = 4.13KK300 pKa = 10.66DD301 pKa = 3.12GDD303 pKa = 4.14NIVSLTIFRR312 pKa = 11.84EE313 pKa = 4.47DD314 pKa = 3.88YY315 pKa = 10.21LTGQSLQLRR324 pKa = 11.84ISSDD328 pKa = 3.03IEE330 pKa = 3.88EE331 pKa = 4.29IEE333 pKa = 4.45INGLTIGMANLLMSMHH349 pKa = 6.36YY350 pKa = 10.6NSGAYY355 pKa = 8.9YY356 pKa = 10.0VAPLDD361 pKa = 4.11AYY363 pKa = 10.99VGLSLARR370 pKa = 11.84LHH372 pKa = 6.18DD373 pKa = 3.74TVYY376 pKa = 11.27NSGPINNGEE385 pKa = 4.26SYY387 pKa = 11.17DD388 pKa = 3.94EE389 pKa = 4.18NLAPTFDD396 pKa = 4.82SNGQSTGVAVSSAIGGTWTTPEE418 pKa = 3.81IATEE422 pKa = 4.2SHH424 pKa = 6.64HH425 pKa = 5.9GWMVTGVTSDD435 pKa = 3.1SGYY438 pKa = 6.97TTLNTTSALKK448 pKa = 10.49EE449 pKa = 3.73IFFAEE454 pKa = 5.65GITAEE459 pKa = 4.39DD460 pKa = 3.51VRR462 pKa = 11.84FTATSSGVDD471 pKa = 3.29NAGLTIHH478 pKa = 7.4IDD480 pKa = 3.21RR481 pKa = 11.84LNYY484 pKa = 9.43SLSVAGFEE492 pKa = 4.64GGRR495 pKa = 11.84TVSGIGFYY503 pKa = 11.02DD504 pKa = 3.74SALHH508 pKa = 5.75AHH510 pKa = 6.32VSSAAAVSLVGTAPGQYY527 pKa = 8.25QVRR530 pKa = 11.84LSGEE534 pKa = 3.84TSYY537 pKa = 11.26TPNAVSATYY546 pKa = 10.09FLEE549 pKa = 4.2ALNFADD555 pKa = 3.88STVIDD560 pKa = 4.98LLNAPLTFKK569 pKa = 10.98GSALNEE575 pKa = 3.97TLYY578 pKa = 11.46GLNTRR583 pKa = 11.84GDD585 pKa = 4.55LIFGLAGNDD594 pKa = 3.52TIYY597 pKa = 10.67GYY599 pKa = 11.25GGADD603 pKa = 3.41VLHH606 pKa = 6.86GGDD609 pKa = 4.41GADD612 pKa = 3.91ALHH615 pKa = 6.81GGDD618 pKa = 5.51GNDD621 pKa = 3.35TLYY624 pKa = 11.45GDD626 pKa = 4.22GGIDD630 pKa = 3.27ALYY633 pKa = 10.42GGEE636 pKa = 4.45GNDD639 pKa = 3.35VLYY642 pKa = 11.19GGDD645 pKa = 4.32GNDD648 pKa = 3.81NLNGEE653 pKa = 4.45NGNDD657 pKa = 3.83TLHH660 pKa = 6.89GGAGNDD666 pKa = 3.47NLRR669 pKa = 11.84GGTGDD674 pKa = 3.44DD675 pKa = 3.46TYY677 pKa = 11.91VFARR681 pKa = 11.84VFSATTAGDD690 pKa = 3.88IIYY693 pKa = 10.34EE694 pKa = 4.07NAADD698 pKa = 4.06GFDD701 pKa = 3.93TIFFTDD707 pKa = 5.12GIEE710 pKa = 4.03ADD712 pKa = 4.46EE713 pKa = 4.63VYY715 pKa = 10.69SWTDD719 pKa = 2.83TSGFLWMQVGGAAATNTLKK738 pKa = 11.07AQGSYY743 pKa = 10.57AAATGITTRR752 pKa = 11.84IEE754 pKa = 3.98QVTFDD759 pKa = 5.32DD760 pKa = 3.79ATVWDD765 pKa = 4.63LTQGLHH771 pKa = 6.52LRR773 pKa = 11.84NNDD776 pKa = 3.29TARR779 pKa = 11.84ALYY782 pKa = 8.02GTIYY786 pKa = 10.89ADD788 pKa = 3.79TIEE791 pKa = 4.92GGSAADD797 pKa = 3.81NIYY800 pKa = 11.1GLGGNDD806 pKa = 3.34TLIGHH811 pKa = 7.42GGNDD815 pKa = 3.69ALRR818 pKa = 11.84GGAGDD823 pKa = 3.68DD824 pKa = 3.54TYY826 pKa = 11.52IFARR830 pKa = 11.84NFSGTTAGDD839 pKa = 3.86IIYY842 pKa = 10.59EE843 pKa = 4.18NVDD846 pKa = 3.5EE847 pKa = 5.03GNDD850 pKa = 3.78TIFFTDD856 pKa = 5.36GIDD859 pKa = 3.44ASEE862 pKa = 4.52VYY864 pKa = 10.25SWTDD868 pKa = 2.81TSGFLWMQVGSAAATNTLKK887 pKa = 11.07AQGSYY892 pKa = 10.87AAVTGITTRR901 pKa = 11.84IEE903 pKa = 3.88QVVFDD908 pKa = 5.87DD909 pKa = 3.73EE910 pKa = 4.98TVWDD914 pKa = 4.24LTQGLHH920 pKa = 6.52LRR922 pKa = 11.84NNDD925 pKa = 3.43TARR928 pKa = 11.84TFYY931 pKa = 9.89GTVYY935 pKa = 11.09GDD937 pKa = 3.59TLEE940 pKa = 4.85GGSASDD946 pKa = 4.27HH947 pKa = 6.57IYY949 pKa = 11.26GLGGNDD955 pKa = 3.34TLIGHH960 pKa = 7.36GGNDD964 pKa = 3.64TLRR967 pKa = 11.84GGDD970 pKa = 3.57GDD972 pKa = 3.91DD973 pKa = 3.5TYY975 pKa = 12.1VFATGFSLNATGDD988 pKa = 4.16RR989 pKa = 11.84IVEE992 pKa = 4.27TATGGTDD999 pKa = 3.43TILFTDD1005 pKa = 5.84GIGADD1010 pKa = 5.57DD1011 pKa = 3.95MFSWTDD1017 pKa = 3.07TSGNLWLQLAANPVSNTLKK1036 pKa = 10.28IDD1038 pKa = 3.65GAYY1041 pKa = 10.01SASTGITTYY1050 pKa = 10.82VEE1052 pKa = 3.95NVVFDD1057 pKa = 5.65DD1058 pKa = 3.53ATAWDD1063 pKa = 4.38LTQGLYY1069 pKa = 10.64LRR1071 pKa = 11.84NNDD1074 pKa = 3.94TGRR1077 pKa = 11.84QMYY1080 pKa = 10.36GSALGDD1086 pKa = 3.86WIEE1089 pKa = 4.57GGAGADD1095 pKa = 3.51TLRR1098 pKa = 11.84GFGGDD1103 pKa = 3.28DD1104 pKa = 3.34TLIGRR1109 pKa = 11.84GGNEE1113 pKa = 3.92DD1114 pKa = 4.11LYY1116 pKa = 11.09GGSGADD1122 pKa = 2.95TFVFRR1127 pKa = 11.84AVDD1130 pKa = 3.4VGNGIDD1136 pKa = 3.59TLRR1139 pKa = 11.84DD1140 pKa = 3.33FSMADD1145 pKa = 3.02NDD1147 pKa = 4.43VIDD1150 pKa = 5.11LRR1152 pKa = 11.84DD1153 pKa = 3.61VLDD1156 pKa = 4.28GGFDD1160 pKa = 3.66PMTDD1164 pKa = 3.18ALSDD1168 pKa = 3.7FVQFTNAGSHH1178 pKa = 4.75SQLRR1182 pKa = 11.84VDD1184 pKa = 4.76LDD1186 pKa = 4.03GAGTLYY1192 pKa = 10.73GWTHH1196 pKa = 6.6IATVNGHH1203 pKa = 5.78TNLDD1207 pKa = 3.64ATTLEE1212 pKa = 4.24ANGYY1216 pKa = 9.43LLAAA1220 pKa = 5.2
Molecular weight: 129.43 kDa
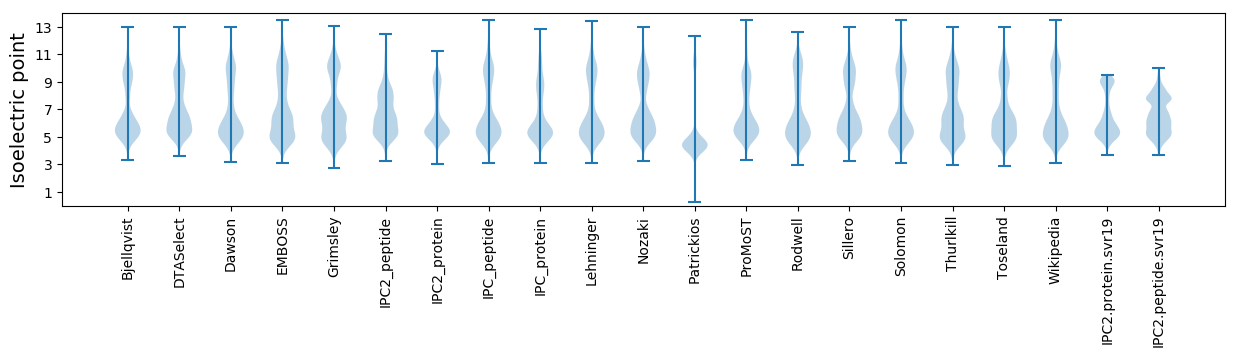
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9YA53|A0A1X9YA53_9SPHN Uncharacterized protein OS=Sphingomonas sp. KC8 OX=1030157 GN=KC8_00090 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.61GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.61GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1230456 |
29 |
4170 |
319.9 |
34.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.852 ± 0.061 | 0.763 ± 0.011 |
6.153 ± 0.03 | 5.061 ± 0.034 |
3.502 ± 0.023 | 9.115 ± 0.057 |
2.026 ± 0.02 | 5.327 ± 0.029 |
2.848 ± 0.027 | 9.667 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.478 ± 0.019 | 2.53 ± 0.031 |
5.31 ± 0.033 | 3.014 ± 0.022 |
7.383 ± 0.046 | 4.974 ± 0.03 |
5.293 ± 0.039 | 7.121 ± 0.032 |
1.39 ± 0.016 | 2.193 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
