
Heterobasidion partitivirus 8
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
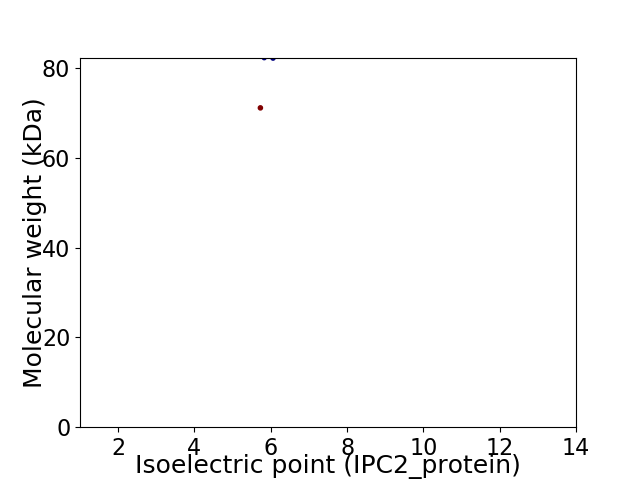
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7U6Q4|K7U6Q4_9VIRU Capsid protein OS=Heterobasidion partitivirus 8 OX=1249677 PE=4 SV=1
MM1 pKa = 7.6ASSATRR7 pKa = 11.84IQTASRR13 pKa = 11.84VSPVEE18 pKa = 3.43IRR20 pKa = 11.84TPPALAAIAKK30 pKa = 9.41KK31 pKa = 9.64FAPVVLQDD39 pKa = 4.88DD40 pKa = 4.44DD41 pKa = 4.61TPVPPSNADD50 pKa = 2.98YY51 pKa = 10.32FGRR54 pKa = 11.84MNLPTNVEE62 pKa = 4.1LDD64 pKa = 3.04SGNYY68 pKa = 9.78HH69 pKa = 6.72NIDD72 pKa = 3.33VQFDD76 pKa = 3.36TRR78 pKa = 11.84FILNYY83 pKa = 10.02IMYY86 pKa = 9.24HH87 pKa = 4.51VTALAPEE94 pKa = 4.72LNFKK98 pKa = 9.16GHH100 pKa = 7.38PYY102 pKa = 10.3FSPLSYY108 pKa = 9.8IGYY111 pKa = 9.38CLHH114 pKa = 7.17LLYY117 pKa = 11.04ALMLACDD124 pKa = 3.41VTFRR128 pKa = 11.84SDD130 pKa = 3.79KK131 pKa = 9.68SWHH134 pKa = 6.24AARR137 pKa = 11.84FMTDD141 pKa = 2.93QEE143 pKa = 4.42RR144 pKa = 11.84KK145 pKa = 9.99DD146 pKa = 3.61LYY148 pKa = 10.82EE149 pKa = 4.03VLLNCHH155 pKa = 6.19MPTFLSDD162 pKa = 4.29LFLEE166 pKa = 4.76LSPVYY171 pKa = 10.33DD172 pKa = 3.59PRR174 pKa = 11.84RR175 pKa = 11.84NNLLFVPSLAGFSFEE190 pKa = 4.7HH191 pKa = 6.97DD192 pKa = 4.04FGRR195 pKa = 11.84TIPPSLFYY203 pKa = 10.53AAHH206 pKa = 6.59HH207 pKa = 6.01MLASTRR213 pKa = 11.84TNKK216 pKa = 10.37DD217 pKa = 3.26PNDD220 pKa = 3.98VIDD223 pKa = 5.15DD224 pKa = 4.02CMSLNVITAGATTYY238 pKa = 10.58KK239 pKa = 10.42VSNFLGTWYY248 pKa = 10.63ASGHH252 pKa = 5.92HH253 pKa = 6.98DD254 pKa = 2.79NWLNRR259 pKa = 11.84DD260 pKa = 3.55FLAFFNPLVGRR271 pKa = 11.84FLTQRR276 pKa = 11.84PTFARR281 pKa = 11.84IHH283 pKa = 6.57TDD285 pKa = 3.08TEE287 pKa = 4.02TLAIDD292 pKa = 3.92GTGNPYY298 pKa = 8.8TAYY301 pKa = 11.09LLASDD306 pKa = 4.73EE307 pKa = 4.42NTSVMTTLLTAMSTFILANDD327 pKa = 4.05PKK329 pKa = 10.63APKK332 pKa = 9.79LGSVLATLSGTLLLSYY348 pKa = 10.33SIEE351 pKa = 4.41PPTLPTWTAATYY363 pKa = 7.69TQEE366 pKa = 3.98TDD368 pKa = 3.24PADD371 pKa = 3.65VNDD374 pKa = 3.46ATFARR379 pKa = 11.84EE380 pKa = 4.1HH381 pKa = 6.42NFLADD386 pKa = 3.58EE387 pKa = 4.2PVYY390 pKa = 10.8TNDD393 pKa = 4.61GEE395 pKa = 4.6YY396 pKa = 10.23PDD398 pKa = 5.15DD399 pKa = 3.75TTNGNMQLYY408 pKa = 9.22HH409 pKa = 6.56FAHH412 pKa = 7.38GAPAHH417 pKa = 5.33TAARR421 pKa = 11.84TPYY424 pKa = 10.21KK425 pKa = 10.43HH426 pKa = 6.54IVFNARR432 pKa = 11.84YY433 pKa = 8.45HH434 pKa = 5.16LTPYY438 pKa = 10.77VMYY441 pKa = 8.66FQPYY445 pKa = 8.83DD446 pKa = 3.64VSPSSLGLTIAAGIKK461 pKa = 9.46IEE463 pKa = 4.62HH464 pKa = 6.84GDD466 pKa = 3.27IAGFAVNIEE475 pKa = 4.18QPEE478 pKa = 4.23SSLDD482 pKa = 3.7DD483 pKa = 3.68NNAQILQSAIRR494 pKa = 11.84LNKK497 pKa = 9.77IIRR500 pKa = 11.84VNANTTAGNNASRR513 pKa = 11.84VVQRR517 pKa = 11.84DD518 pKa = 3.04HH519 pKa = 7.9LDD521 pKa = 3.32RR522 pKa = 11.84TKK524 pKa = 10.85QGIMTIFRR532 pKa = 11.84SIAKK536 pKa = 9.96SVFPYY541 pKa = 10.73LDD543 pKa = 3.35GSAIHH548 pKa = 5.65YY549 pKa = 7.7TPATLPRR556 pKa = 11.84NLGLTEE562 pKa = 4.04EE563 pKa = 4.48AGHH566 pKa = 6.55YY567 pKa = 10.34SLQQGFNVKK576 pKa = 10.28AGTEE580 pKa = 4.36GKK582 pKa = 8.46FTAPDD587 pKa = 3.23HH588 pKa = 7.26SIYY591 pKa = 10.55LWSSYY596 pKa = 10.72RR597 pKa = 11.84YY598 pKa = 8.02VHH600 pKa = 6.49KK601 pKa = 10.6KK602 pKa = 10.09KK603 pKa = 10.65NPAPADD609 pKa = 3.15ISMLATLRR617 pKa = 11.84TFYY620 pKa = 10.97GSNVTLSRR628 pKa = 11.84SKK630 pKa = 11.16NPVLMIPHH638 pKa = 7.3
MM1 pKa = 7.6ASSATRR7 pKa = 11.84IQTASRR13 pKa = 11.84VSPVEE18 pKa = 3.43IRR20 pKa = 11.84TPPALAAIAKK30 pKa = 9.41KK31 pKa = 9.64FAPVVLQDD39 pKa = 4.88DD40 pKa = 4.44DD41 pKa = 4.61TPVPPSNADD50 pKa = 2.98YY51 pKa = 10.32FGRR54 pKa = 11.84MNLPTNVEE62 pKa = 4.1LDD64 pKa = 3.04SGNYY68 pKa = 9.78HH69 pKa = 6.72NIDD72 pKa = 3.33VQFDD76 pKa = 3.36TRR78 pKa = 11.84FILNYY83 pKa = 10.02IMYY86 pKa = 9.24HH87 pKa = 4.51VTALAPEE94 pKa = 4.72LNFKK98 pKa = 9.16GHH100 pKa = 7.38PYY102 pKa = 10.3FSPLSYY108 pKa = 9.8IGYY111 pKa = 9.38CLHH114 pKa = 7.17LLYY117 pKa = 11.04ALMLACDD124 pKa = 3.41VTFRR128 pKa = 11.84SDD130 pKa = 3.79KK131 pKa = 9.68SWHH134 pKa = 6.24AARR137 pKa = 11.84FMTDD141 pKa = 2.93QEE143 pKa = 4.42RR144 pKa = 11.84KK145 pKa = 9.99DD146 pKa = 3.61LYY148 pKa = 10.82EE149 pKa = 4.03VLLNCHH155 pKa = 6.19MPTFLSDD162 pKa = 4.29LFLEE166 pKa = 4.76LSPVYY171 pKa = 10.33DD172 pKa = 3.59PRR174 pKa = 11.84RR175 pKa = 11.84NNLLFVPSLAGFSFEE190 pKa = 4.7HH191 pKa = 6.97DD192 pKa = 4.04FGRR195 pKa = 11.84TIPPSLFYY203 pKa = 10.53AAHH206 pKa = 6.59HH207 pKa = 6.01MLASTRR213 pKa = 11.84TNKK216 pKa = 10.37DD217 pKa = 3.26PNDD220 pKa = 3.98VIDD223 pKa = 5.15DD224 pKa = 4.02CMSLNVITAGATTYY238 pKa = 10.58KK239 pKa = 10.42VSNFLGTWYY248 pKa = 10.63ASGHH252 pKa = 5.92HH253 pKa = 6.98DD254 pKa = 2.79NWLNRR259 pKa = 11.84DD260 pKa = 3.55FLAFFNPLVGRR271 pKa = 11.84FLTQRR276 pKa = 11.84PTFARR281 pKa = 11.84IHH283 pKa = 6.57TDD285 pKa = 3.08TEE287 pKa = 4.02TLAIDD292 pKa = 3.92GTGNPYY298 pKa = 8.8TAYY301 pKa = 11.09LLASDD306 pKa = 4.73EE307 pKa = 4.42NTSVMTTLLTAMSTFILANDD327 pKa = 4.05PKK329 pKa = 10.63APKK332 pKa = 9.79LGSVLATLSGTLLLSYY348 pKa = 10.33SIEE351 pKa = 4.41PPTLPTWTAATYY363 pKa = 7.69TQEE366 pKa = 3.98TDD368 pKa = 3.24PADD371 pKa = 3.65VNDD374 pKa = 3.46ATFARR379 pKa = 11.84EE380 pKa = 4.1HH381 pKa = 6.42NFLADD386 pKa = 3.58EE387 pKa = 4.2PVYY390 pKa = 10.8TNDD393 pKa = 4.61GEE395 pKa = 4.6YY396 pKa = 10.23PDD398 pKa = 5.15DD399 pKa = 3.75TTNGNMQLYY408 pKa = 9.22HH409 pKa = 6.56FAHH412 pKa = 7.38GAPAHH417 pKa = 5.33TAARR421 pKa = 11.84TPYY424 pKa = 10.21KK425 pKa = 10.43HH426 pKa = 6.54IVFNARR432 pKa = 11.84YY433 pKa = 8.45HH434 pKa = 5.16LTPYY438 pKa = 10.77VMYY441 pKa = 8.66FQPYY445 pKa = 8.83DD446 pKa = 3.64VSPSSLGLTIAAGIKK461 pKa = 9.46IEE463 pKa = 4.62HH464 pKa = 6.84GDD466 pKa = 3.27IAGFAVNIEE475 pKa = 4.18QPEE478 pKa = 4.23SSLDD482 pKa = 3.7DD483 pKa = 3.68NNAQILQSAIRR494 pKa = 11.84LNKK497 pKa = 9.77IIRR500 pKa = 11.84VNANTTAGNNASRR513 pKa = 11.84VVQRR517 pKa = 11.84DD518 pKa = 3.04HH519 pKa = 7.9LDD521 pKa = 3.32RR522 pKa = 11.84TKK524 pKa = 10.85QGIMTIFRR532 pKa = 11.84SIAKK536 pKa = 9.96SVFPYY541 pKa = 10.73LDD543 pKa = 3.35GSAIHH548 pKa = 5.65YY549 pKa = 7.7TPATLPRR556 pKa = 11.84NLGLTEE562 pKa = 4.04EE563 pKa = 4.48AGHH566 pKa = 6.55YY567 pKa = 10.34SLQQGFNVKK576 pKa = 10.28AGTEE580 pKa = 4.36GKK582 pKa = 8.46FTAPDD587 pKa = 3.23HH588 pKa = 7.26SIYY591 pKa = 10.55LWSSYY596 pKa = 10.72RR597 pKa = 11.84YY598 pKa = 8.02VHH600 pKa = 6.49KK601 pKa = 10.6KK602 pKa = 10.09KK603 pKa = 10.65NPAPADD609 pKa = 3.15ISMLATLRR617 pKa = 11.84TFYY620 pKa = 10.97GSNVTLSRR628 pKa = 11.84SKK630 pKa = 11.16NPVLMIPHH638 pKa = 7.3
Molecular weight: 71.11 kDa
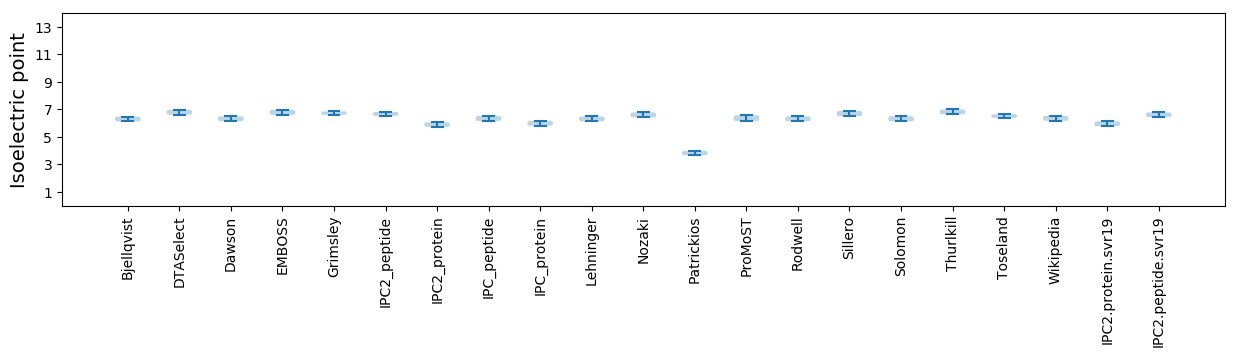
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7U6Q4|K7U6Q4_9VIRU Capsid protein OS=Heterobasidion partitivirus 8 OX=1249677 PE=4 SV=1
MM1 pKa = 7.39LSKK4 pKa = 10.76VRR6 pKa = 11.84DD7 pKa = 3.98FFHH10 pKa = 7.12EE11 pKa = 4.08KK12 pKa = 10.4LSRR15 pKa = 11.84LLLQHH20 pKa = 7.19SIFQSNDD27 pKa = 2.54KK28 pKa = 11.21DD29 pKa = 3.79EE30 pKa = 4.45EE31 pKa = 4.22QTLLEE36 pKa = 4.36NQSSDD41 pKa = 2.88VKK43 pKa = 10.59RR44 pKa = 11.84IYY46 pKa = 11.07NSIHH50 pKa = 6.42YY51 pKa = 9.74SFQTDD56 pKa = 3.48DD57 pKa = 3.26TQAAYY62 pKa = 10.03EE63 pKa = 4.37DD64 pKa = 4.24QYY66 pKa = 11.56QHH68 pKa = 6.37IKK70 pKa = 10.44HH71 pKa = 5.68VLEE74 pKa = 5.12DD75 pKa = 3.85KK76 pKa = 10.95DD77 pKa = 3.76RR78 pKa = 11.84LSNFPAEE85 pKa = 4.76FYY87 pKa = 10.96LPYY90 pKa = 10.06DD91 pKa = 3.62TTTTPDD97 pKa = 3.0NRR99 pKa = 11.84VPPSGIDD106 pKa = 3.24QLPYY110 pKa = 9.75VYY112 pKa = 10.66KK113 pKa = 9.56RR114 pKa = 11.84TNVVTATDD122 pKa = 3.91EE123 pKa = 4.35VPEE126 pKa = 4.06TGYY129 pKa = 9.95PIQNRR134 pKa = 11.84LLRR137 pKa = 11.84LIRR140 pKa = 11.84SRR142 pKa = 11.84YY143 pKa = 7.28PQYY146 pKa = 10.62LPHH149 pKa = 6.38VRR151 pKa = 11.84TFTRR155 pKa = 11.84PLGTTDD161 pKa = 2.98ATVSDD166 pKa = 4.67FFKK169 pKa = 10.07PQHH172 pKa = 6.49PSRR175 pKa = 11.84LVDD178 pKa = 3.71PSRR181 pKa = 11.84ISHH184 pKa = 5.13VMKK187 pKa = 10.62HH188 pKa = 5.76VMNKK192 pKa = 8.36MAITPYY198 pKa = 10.69LPIHH202 pKa = 6.26FVDD205 pKa = 3.47TQYY208 pKa = 11.77DD209 pKa = 3.79KK210 pKa = 11.46RR211 pKa = 11.84PLSTGTGYY219 pKa = 10.91YY220 pKa = 10.02NRR222 pKa = 11.84RR223 pKa = 11.84SHH225 pKa = 6.34EE226 pKa = 4.28ANIHH230 pKa = 5.7ALYY233 pKa = 10.35SHH235 pKa = 7.03PKK237 pKa = 8.41EE238 pKa = 4.3YY239 pKa = 10.69EE240 pKa = 3.71NKK242 pKa = 8.33RR243 pKa = 11.84TSKK246 pKa = 10.61GYY248 pKa = 10.13YY249 pKa = 8.92INAFLEE255 pKa = 4.47SARR258 pKa = 11.84SLVHH262 pKa = 6.62WIKK265 pKa = 11.0SYY267 pKa = 11.51GNPFRR272 pKa = 11.84SKK274 pKa = 10.12PADD277 pKa = 3.33PRR279 pKa = 11.84EE280 pKa = 4.08SLKK283 pKa = 10.94KK284 pKa = 10.52FFLQRR289 pKa = 11.84PTMLFTRR296 pKa = 11.84NHH298 pKa = 5.57ISKK301 pKa = 10.34ILGALKK307 pKa = 9.86QRR309 pKa = 11.84PVYY312 pKa = 10.64AVDD315 pKa = 5.16DD316 pKa = 4.18LFLTLEE322 pKa = 4.22SMVTFPAHH330 pKa = 5.7TIARR334 pKa = 11.84KK335 pKa = 9.53IEE337 pKa = 3.89CCIMYY342 pKa = 9.9GYY344 pKa = 8.32EE345 pKa = 4.2TIRR348 pKa = 11.84GSNVQLDD355 pKa = 4.04RR356 pKa = 11.84LAQRR360 pKa = 11.84YY361 pKa = 8.96NSFFTIDD368 pKa = 2.88WSGFDD373 pKa = 4.51QRR375 pKa = 11.84LPWVIVLLFFTEE387 pKa = 4.43FLPRR391 pKa = 11.84LLVINHH397 pKa = 6.91GYY399 pKa = 10.56APTYY403 pKa = 9.87DD404 pKa = 3.63FPSYY408 pKa = 10.67PDD410 pKa = 3.43LTTEE414 pKa = 3.76MMYY417 pKa = 10.55QRR419 pKa = 11.84LSNILSFLATWYY431 pKa = 10.65FNMVFITADD440 pKa = 2.7GFAYY444 pKa = 9.87VRR446 pKa = 11.84RR447 pKa = 11.84FAGVPSGLLNTQFLDD462 pKa = 3.52SFGNLFLIIDD472 pKa = 3.84ALIEE476 pKa = 4.05FGAQDD481 pKa = 3.8EE482 pKa = 5.07EE483 pKa = 4.5IDD485 pKa = 4.79SILLLIMGDD494 pKa = 3.84DD495 pKa = 3.68NSGFTIWSIARR506 pKa = 11.84LEE508 pKa = 3.93QFITFLEE515 pKa = 4.99SYY517 pKa = 10.76ALTRR521 pKa = 11.84YY522 pKa = 9.48GMVMSKK528 pKa = 9.09TKK530 pKa = 10.84SLVTVLRR537 pKa = 11.84HH538 pKa = 6.49KK539 pKa = 10.52IQTLSYY545 pKa = 8.4TCNFGRR551 pKa = 11.84PLRR554 pKa = 11.84PIPEE558 pKa = 4.38LVAHH562 pKa = 6.37LVFPEE567 pKa = 4.63RR568 pKa = 11.84EE569 pKa = 4.1FKK571 pKa = 10.1PQFMSARR578 pKa = 11.84AVGMAWASCAQDD590 pKa = 3.22KK591 pKa = 9.26TFHH594 pKa = 6.69DD595 pKa = 4.71FCRR598 pKa = 11.84DD599 pKa = 2.86VFYY602 pKa = 10.52EE603 pKa = 3.96YY604 pKa = 10.92LEE606 pKa = 4.25EE607 pKa = 4.66SVPVDD612 pKa = 3.3NTNIAWIQSHH622 pKa = 6.37LPGYY626 pKa = 10.34LRR628 pKa = 11.84VDD630 pKa = 3.52PEE632 pKa = 4.16VTKK635 pKa = 10.55MIDD638 pKa = 3.39LNVFPSFLHH647 pKa = 6.08VSQKK651 pKa = 10.14LSRR654 pKa = 11.84WQGPLSYY661 pKa = 10.28QPKK664 pKa = 8.69WDD666 pKa = 3.58LAHH669 pKa = 7.45FINQPDD675 pKa = 4.46VIPDD679 pKa = 3.44DD680 pKa = 4.7SITMFEE686 pKa = 4.06YY687 pKa = 9.82MQEE690 pKa = 4.03HH691 pKa = 6.57SLSLDD696 pKa = 3.06IQFDD700 pKa = 3.97LFSAA704 pKa = 4.31
MM1 pKa = 7.39LSKK4 pKa = 10.76VRR6 pKa = 11.84DD7 pKa = 3.98FFHH10 pKa = 7.12EE11 pKa = 4.08KK12 pKa = 10.4LSRR15 pKa = 11.84LLLQHH20 pKa = 7.19SIFQSNDD27 pKa = 2.54KK28 pKa = 11.21DD29 pKa = 3.79EE30 pKa = 4.45EE31 pKa = 4.22QTLLEE36 pKa = 4.36NQSSDD41 pKa = 2.88VKK43 pKa = 10.59RR44 pKa = 11.84IYY46 pKa = 11.07NSIHH50 pKa = 6.42YY51 pKa = 9.74SFQTDD56 pKa = 3.48DD57 pKa = 3.26TQAAYY62 pKa = 10.03EE63 pKa = 4.37DD64 pKa = 4.24QYY66 pKa = 11.56QHH68 pKa = 6.37IKK70 pKa = 10.44HH71 pKa = 5.68VLEE74 pKa = 5.12DD75 pKa = 3.85KK76 pKa = 10.95DD77 pKa = 3.76RR78 pKa = 11.84LSNFPAEE85 pKa = 4.76FYY87 pKa = 10.96LPYY90 pKa = 10.06DD91 pKa = 3.62TTTTPDD97 pKa = 3.0NRR99 pKa = 11.84VPPSGIDD106 pKa = 3.24QLPYY110 pKa = 9.75VYY112 pKa = 10.66KK113 pKa = 9.56RR114 pKa = 11.84TNVVTATDD122 pKa = 3.91EE123 pKa = 4.35VPEE126 pKa = 4.06TGYY129 pKa = 9.95PIQNRR134 pKa = 11.84LLRR137 pKa = 11.84LIRR140 pKa = 11.84SRR142 pKa = 11.84YY143 pKa = 7.28PQYY146 pKa = 10.62LPHH149 pKa = 6.38VRR151 pKa = 11.84TFTRR155 pKa = 11.84PLGTTDD161 pKa = 2.98ATVSDD166 pKa = 4.67FFKK169 pKa = 10.07PQHH172 pKa = 6.49PSRR175 pKa = 11.84LVDD178 pKa = 3.71PSRR181 pKa = 11.84ISHH184 pKa = 5.13VMKK187 pKa = 10.62HH188 pKa = 5.76VMNKK192 pKa = 8.36MAITPYY198 pKa = 10.69LPIHH202 pKa = 6.26FVDD205 pKa = 3.47TQYY208 pKa = 11.77DD209 pKa = 3.79KK210 pKa = 11.46RR211 pKa = 11.84PLSTGTGYY219 pKa = 10.91YY220 pKa = 10.02NRR222 pKa = 11.84RR223 pKa = 11.84SHH225 pKa = 6.34EE226 pKa = 4.28ANIHH230 pKa = 5.7ALYY233 pKa = 10.35SHH235 pKa = 7.03PKK237 pKa = 8.41EE238 pKa = 4.3YY239 pKa = 10.69EE240 pKa = 3.71NKK242 pKa = 8.33RR243 pKa = 11.84TSKK246 pKa = 10.61GYY248 pKa = 10.13YY249 pKa = 8.92INAFLEE255 pKa = 4.47SARR258 pKa = 11.84SLVHH262 pKa = 6.62WIKK265 pKa = 11.0SYY267 pKa = 11.51GNPFRR272 pKa = 11.84SKK274 pKa = 10.12PADD277 pKa = 3.33PRR279 pKa = 11.84EE280 pKa = 4.08SLKK283 pKa = 10.94KK284 pKa = 10.52FFLQRR289 pKa = 11.84PTMLFTRR296 pKa = 11.84NHH298 pKa = 5.57ISKK301 pKa = 10.34ILGALKK307 pKa = 9.86QRR309 pKa = 11.84PVYY312 pKa = 10.64AVDD315 pKa = 5.16DD316 pKa = 4.18LFLTLEE322 pKa = 4.22SMVTFPAHH330 pKa = 5.7TIARR334 pKa = 11.84KK335 pKa = 9.53IEE337 pKa = 3.89CCIMYY342 pKa = 9.9GYY344 pKa = 8.32EE345 pKa = 4.2TIRR348 pKa = 11.84GSNVQLDD355 pKa = 4.04RR356 pKa = 11.84LAQRR360 pKa = 11.84YY361 pKa = 8.96NSFFTIDD368 pKa = 2.88WSGFDD373 pKa = 4.51QRR375 pKa = 11.84LPWVIVLLFFTEE387 pKa = 4.43FLPRR391 pKa = 11.84LLVINHH397 pKa = 6.91GYY399 pKa = 10.56APTYY403 pKa = 9.87DD404 pKa = 3.63FPSYY408 pKa = 10.67PDD410 pKa = 3.43LTTEE414 pKa = 3.76MMYY417 pKa = 10.55QRR419 pKa = 11.84LSNILSFLATWYY431 pKa = 10.65FNMVFITADD440 pKa = 2.7GFAYY444 pKa = 9.87VRR446 pKa = 11.84RR447 pKa = 11.84FAGVPSGLLNTQFLDD462 pKa = 3.52SFGNLFLIIDD472 pKa = 3.84ALIEE476 pKa = 4.05FGAQDD481 pKa = 3.8EE482 pKa = 5.07EE483 pKa = 4.5IDD485 pKa = 4.79SILLLIMGDD494 pKa = 3.84DD495 pKa = 3.68NSGFTIWSIARR506 pKa = 11.84LEE508 pKa = 3.93QFITFLEE515 pKa = 4.99SYY517 pKa = 10.76ALTRR521 pKa = 11.84YY522 pKa = 9.48GMVMSKK528 pKa = 9.09TKK530 pKa = 10.84SLVTVLRR537 pKa = 11.84HH538 pKa = 6.49KK539 pKa = 10.52IQTLSYY545 pKa = 8.4TCNFGRR551 pKa = 11.84PLRR554 pKa = 11.84PIPEE558 pKa = 4.38LVAHH562 pKa = 6.37LVFPEE567 pKa = 4.63RR568 pKa = 11.84EE569 pKa = 4.1FKK571 pKa = 10.1PQFMSARR578 pKa = 11.84AVGMAWASCAQDD590 pKa = 3.22KK591 pKa = 9.26TFHH594 pKa = 6.69DD595 pKa = 4.71FCRR598 pKa = 11.84DD599 pKa = 2.86VFYY602 pKa = 10.52EE603 pKa = 3.96YY604 pKa = 10.92LEE606 pKa = 4.25EE607 pKa = 4.66SVPVDD612 pKa = 3.3NTNIAWIQSHH622 pKa = 6.37LPGYY626 pKa = 10.34LRR628 pKa = 11.84VDD630 pKa = 3.52PEE632 pKa = 4.16VTKK635 pKa = 10.55MIDD638 pKa = 3.39LNVFPSFLHH647 pKa = 6.08VSQKK651 pKa = 10.14LSRR654 pKa = 11.84WQGPLSYY661 pKa = 10.28QPKK664 pKa = 8.69WDD666 pKa = 3.58LAHH669 pKa = 7.45FINQPDD675 pKa = 4.46VIPDD679 pKa = 3.44DD680 pKa = 4.7SITMFEE686 pKa = 4.06YY687 pKa = 9.82MQEE690 pKa = 4.03HH691 pKa = 6.57SLSLDD696 pKa = 3.06IQFDD700 pKa = 3.97LFSAA704 pKa = 4.31
Molecular weight: 82.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1342 |
638 |
704 |
671.0 |
76.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.079 ± 1.707 | 0.671 ± 0.03 |
6.334 ± 0.044 | 3.949 ± 0.56 |
6.11 ± 0.645 | 4.024 ± 0.467 |
3.651 ± 0.184 | 5.44 ± 0.399 |
3.651 ± 0.355 | 9.985 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.459 ± 0.074 | 4.918 ± 0.822 |
6.408 ± 0.228 | 3.502 ± 0.792 |
5.365 ± 0.564 | 7.228 ± 0.228 |
7.675 ± 0.866 | 5.291 ± 0.189 |
1.043 ± 0.178 | 5.216 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
