
Colletotrichum higginsianum non-segmented dsRNA virus 1
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
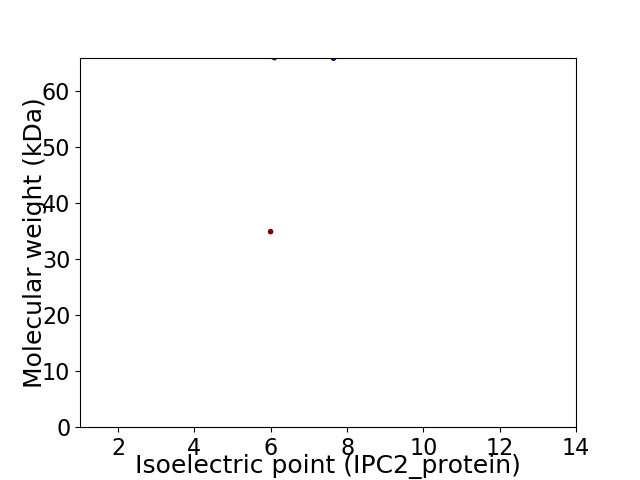
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N7A5G0|A0A0N7A5G0_9VIRU RNA-dependent RNA polymerase OS=Colletotrichum higginsianum non-segmented dsRNA virus 1 OX=1565088 PE=4 SV=1
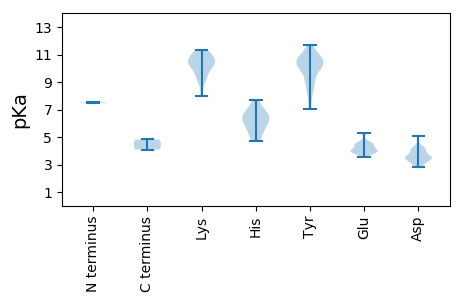
MM1 pKa = 7.52SAPKK5 pKa = 9.91PGASAEE11 pKa = 4.11AKK13 pKa = 9.55PVAGSLPAAGTPAVKK28 pKa = 9.18GTQSKK33 pKa = 10.12EE34 pKa = 4.2SPPAPAPLLGAVAEE48 pKa = 4.32EE49 pKa = 4.15QDD51 pKa = 4.27FVSAPQTPPPLDD63 pKa = 3.3PTRR66 pKa = 11.84FTLSEE71 pKa = 3.67NYY73 pKa = 10.01ARR75 pKa = 11.84LFCEE79 pKa = 3.96RR80 pKa = 11.84NSRR83 pKa = 11.84LAIGDD88 pKa = 4.06IIRR91 pKa = 11.84RR92 pKa = 11.84WLLSKK97 pKa = 11.11GITQGPATPASIYY110 pKa = 9.69PPGWRR115 pKa = 11.84VGRR118 pKa = 11.84TPIPVEE124 pKa = 3.98GGGVGVRR131 pKa = 11.84TLDD134 pKa = 3.24EE135 pKa = 4.11VLPNLRR141 pKa = 11.84AYY143 pKa = 10.63DD144 pKa = 3.7EE145 pKa = 4.4VGNEE149 pKa = 3.95VFVVQPEE156 pKa = 4.54GNWTRR161 pKa = 11.84VARR164 pKa = 11.84LVAVWPAPVTIVAGMLSALQDD185 pKa = 3.51TRR187 pKa = 11.84VTTDD191 pKa = 2.84RR192 pKa = 11.84VLGLAQRR199 pKa = 11.84SPEE202 pKa = 3.88ALEE205 pKa = 3.87KK206 pKa = 10.89LKK208 pKa = 11.04AAGAEE213 pKa = 4.6FKK215 pKa = 11.11AHH217 pKa = 5.02VQRR220 pKa = 11.84FLDD223 pKa = 4.02PVALRR228 pKa = 11.84RR229 pKa = 11.84SPEE232 pKa = 3.87FVEE235 pKa = 4.63VSQRR239 pKa = 11.84YY240 pKa = 8.9KK241 pKa = 10.72RR242 pKa = 11.84EE243 pKa = 3.58LRR245 pKa = 11.84ALIDD249 pKa = 3.5EE250 pKa = 4.24QQRR253 pKa = 11.84AIRR256 pKa = 11.84EE257 pKa = 3.89ARR259 pKa = 11.84AATRR263 pKa = 11.84RR264 pKa = 11.84VNQLLGEE271 pKa = 4.34RR272 pKa = 11.84DD273 pKa = 3.24QALARR278 pKa = 11.84LDD280 pKa = 3.48PAYY283 pKa = 10.64VPKK286 pKa = 10.68RR287 pKa = 11.84ATAAAALAQYY297 pKa = 10.58GIDD300 pKa = 3.43LGEE303 pKa = 4.33EE304 pKa = 4.56GEE306 pKa = 4.49VAGLEE311 pKa = 4.08EE312 pKa = 5.29SEE314 pKa = 4.46TADD317 pKa = 3.95VLATLDD323 pKa = 3.65FF324 pKa = 4.82
MM1 pKa = 7.52SAPKK5 pKa = 9.91PGASAEE11 pKa = 4.11AKK13 pKa = 9.55PVAGSLPAAGTPAVKK28 pKa = 9.18GTQSKK33 pKa = 10.12EE34 pKa = 4.2SPPAPAPLLGAVAEE48 pKa = 4.32EE49 pKa = 4.15QDD51 pKa = 4.27FVSAPQTPPPLDD63 pKa = 3.3PTRR66 pKa = 11.84FTLSEE71 pKa = 3.67NYY73 pKa = 10.01ARR75 pKa = 11.84LFCEE79 pKa = 3.96RR80 pKa = 11.84NSRR83 pKa = 11.84LAIGDD88 pKa = 4.06IIRR91 pKa = 11.84RR92 pKa = 11.84WLLSKK97 pKa = 11.11GITQGPATPASIYY110 pKa = 9.69PPGWRR115 pKa = 11.84VGRR118 pKa = 11.84TPIPVEE124 pKa = 3.98GGGVGVRR131 pKa = 11.84TLDD134 pKa = 3.24EE135 pKa = 4.11VLPNLRR141 pKa = 11.84AYY143 pKa = 10.63DD144 pKa = 3.7EE145 pKa = 4.4VGNEE149 pKa = 3.95VFVVQPEE156 pKa = 4.54GNWTRR161 pKa = 11.84VARR164 pKa = 11.84LVAVWPAPVTIVAGMLSALQDD185 pKa = 3.51TRR187 pKa = 11.84VTTDD191 pKa = 2.84RR192 pKa = 11.84VLGLAQRR199 pKa = 11.84SPEE202 pKa = 3.88ALEE205 pKa = 3.87KK206 pKa = 10.89LKK208 pKa = 11.04AAGAEE213 pKa = 4.6FKK215 pKa = 11.11AHH217 pKa = 5.02VQRR220 pKa = 11.84FLDD223 pKa = 4.02PVALRR228 pKa = 11.84RR229 pKa = 11.84SPEE232 pKa = 3.87FVEE235 pKa = 4.63VSQRR239 pKa = 11.84YY240 pKa = 8.9KK241 pKa = 10.72RR242 pKa = 11.84EE243 pKa = 3.58LRR245 pKa = 11.84ALIDD249 pKa = 3.5EE250 pKa = 4.24QQRR253 pKa = 11.84AIRR256 pKa = 11.84EE257 pKa = 3.89ARR259 pKa = 11.84AATRR263 pKa = 11.84RR264 pKa = 11.84VNQLLGEE271 pKa = 4.34RR272 pKa = 11.84DD273 pKa = 3.24QALARR278 pKa = 11.84LDD280 pKa = 3.48PAYY283 pKa = 10.64VPKK286 pKa = 10.68RR287 pKa = 11.84ATAAAALAQYY297 pKa = 10.58GIDD300 pKa = 3.43LGEE303 pKa = 4.33EE304 pKa = 4.56GEE306 pKa = 4.49VAGLEE311 pKa = 4.08EE312 pKa = 5.29SEE314 pKa = 4.46TADD317 pKa = 3.95VLATLDD323 pKa = 3.65FF324 pKa = 4.82
Molecular weight: 34.86 kDa
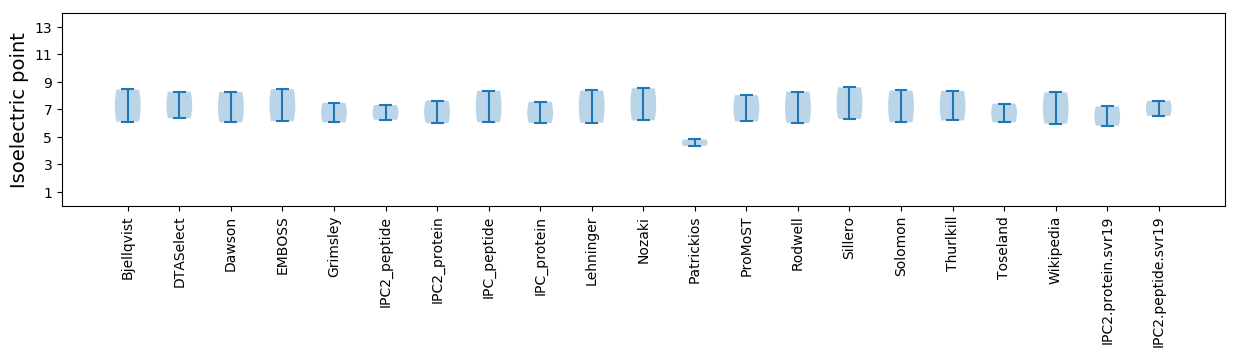
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N7A5G0|A0A0N7A5G0_9VIRU RNA-dependent RNA polymerase OS=Colletotrichum higginsianum non-segmented dsRNA virus 1 OX=1565088 PE=4 SV=1
MM1 pKa = 7.46TEE3 pKa = 3.68ASGLGVKK10 pKa = 9.74DD11 pKa = 2.9WSFFLEE17 pKa = 3.97PAFKK21 pKa = 10.46SKK23 pKa = 9.32EE24 pKa = 3.96LRR26 pKa = 11.84GVRR29 pKa = 11.84CLGFSKK35 pKa = 10.48GLKK38 pKa = 8.88YY39 pKa = 10.58KK40 pKa = 10.44YY41 pKa = 9.73VQPQTKK47 pKa = 9.72NSQFFLSFLDD57 pKa = 3.38QCTMEE62 pKa = 4.51LAGIPGPSLEE72 pKa = 4.41ITTSEE77 pKa = 4.12YY78 pKa = 11.04VPPSEE83 pKa = 4.67SDD85 pKa = 3.41LFAHH89 pKa = 6.94IAGFGEE95 pKa = 4.6SGSSGSQLSNRR106 pKa = 11.84VYY108 pKa = 10.25AALRR112 pKa = 11.84EE113 pKa = 4.12EE114 pKa = 4.44LTMIEE119 pKa = 3.83HH120 pKa = 7.73WDD122 pKa = 3.72RR123 pKa = 11.84EE124 pKa = 4.25ITGWLDD130 pKa = 2.96PRR132 pKa = 11.84LLVDD136 pKa = 4.32IKK138 pKa = 11.23VPSQTSPGIRR148 pKa = 11.84WKK150 pKa = 10.87KK151 pKa = 10.3LGYY154 pKa = 8.14KK155 pKa = 7.97TKK157 pKa = 10.78KK158 pKa = 8.32EE159 pKa = 4.08ALMPATMEE167 pKa = 4.03AAKK170 pKa = 10.57VLLSMIEE177 pKa = 3.88TGTEE181 pKa = 4.23YY182 pKa = 10.41EE183 pKa = 4.34VPPCGVAGRR192 pKa = 11.84GKK194 pKa = 10.61RR195 pKa = 11.84MDD197 pKa = 3.56MNRR200 pKa = 11.84KK201 pKa = 9.27VDD203 pKa = 3.44TGGKK207 pKa = 9.41KK208 pKa = 9.3EE209 pKa = 3.84GRR211 pKa = 11.84LIVMPDD217 pKa = 3.54LVRR220 pKa = 11.84HH221 pKa = 5.67LVGTLGSGAYY231 pKa = 7.84MSKK234 pKa = 10.36VKK236 pKa = 10.44EE237 pKa = 3.97VDD239 pKa = 3.04KK240 pKa = 11.27SQGGILLGMGPFSEE254 pKa = 5.18SYY256 pKa = 10.74QSIANWARR264 pKa = 11.84GASKK268 pKa = 10.2FVFLDD273 pKa = 4.22FKK275 pKa = 11.23KK276 pKa = 10.43FDD278 pKa = 3.43HH279 pKa = 6.42RR280 pKa = 11.84VPARR284 pKa = 11.84VLQEE288 pKa = 4.04VFKK291 pKa = 10.68HH292 pKa = 4.7IASRR296 pKa = 11.84YY297 pKa = 7.05EE298 pKa = 3.78GCPGSKK304 pKa = 10.13AYY306 pKa = 8.89WRR308 pKa = 11.84SEE310 pKa = 3.56FQQLVNTKK318 pKa = 9.77IAMPDD323 pKa = 3.03GTVYY327 pKa = 10.06QKK329 pKa = 10.96KK330 pKa = 10.09RR331 pKa = 11.84GVASGDD337 pKa = 3.07PWTSIAGSYY346 pKa = 9.19ANWMMLKK353 pKa = 10.06HH354 pKa = 5.46VCNVLGLVAKK364 pKa = 10.15IWTFGDD370 pKa = 3.47DD371 pKa = 4.1SVIALYY377 pKa = 10.7DD378 pKa = 3.46VDD380 pKa = 4.23PTLDD384 pKa = 4.07LLPLFTKK391 pKa = 10.52HH392 pKa = 5.95LASSFGMHH400 pKa = 6.18VSQEE404 pKa = 3.66KK405 pKa = 10.75SYY407 pKa = 11.71VSDD410 pKa = 3.85CLVDD414 pKa = 3.28IAEE417 pKa = 4.55DD418 pKa = 4.14PEE420 pKa = 4.63PKK422 pKa = 10.25SSGSFLSLYY431 pKa = 10.3FLQTPMGVRR440 pKa = 11.84PTRR443 pKa = 11.84PLQDD447 pKa = 3.56MYY449 pKa = 11.6EE450 pKa = 3.96MMMVPEE456 pKa = 4.77RR457 pKa = 11.84NQGTLRR463 pKa = 11.84WEE465 pKa = 4.09IVRR468 pKa = 11.84TSMAYY473 pKa = 8.65LTFYY477 pKa = 11.04YY478 pKa = 10.23NDD480 pKa = 3.45RR481 pKa = 11.84ARR483 pKa = 11.84YY484 pKa = 8.91VLEE487 pKa = 5.15EE488 pKa = 3.97YY489 pKa = 10.1WDD491 pKa = 3.58WLHH494 pKa = 6.83RR495 pKa = 11.84KK496 pKa = 9.01YY497 pKa = 10.39RR498 pKa = 11.84VPEE501 pKa = 3.87LKK503 pKa = 11.06GNFTDD508 pKa = 5.06LNLLRR513 pKa = 11.84EE514 pKa = 4.05MDD516 pKa = 4.02IPWSSFKK523 pKa = 11.3LEE525 pKa = 3.88WLVRR529 pKa = 11.84LPRR532 pKa = 11.84PGEE535 pKa = 3.85VEE537 pKa = 4.5LMYY540 pKa = 10.61KK541 pKa = 10.19YY542 pKa = 10.78GHH544 pKa = 6.54TGFFPPLLWGHH555 pKa = 6.64WYY557 pKa = 10.37ARR559 pKa = 11.84YY560 pKa = 10.17DD561 pKa = 3.89RR562 pKa = 11.84DD563 pKa = 3.56VHH565 pKa = 6.22GNNVSFAFQEE575 pKa = 4.46PPTT578 pKa = 4.09
MM1 pKa = 7.46TEE3 pKa = 3.68ASGLGVKK10 pKa = 9.74DD11 pKa = 2.9WSFFLEE17 pKa = 3.97PAFKK21 pKa = 10.46SKK23 pKa = 9.32EE24 pKa = 3.96LRR26 pKa = 11.84GVRR29 pKa = 11.84CLGFSKK35 pKa = 10.48GLKK38 pKa = 8.88YY39 pKa = 10.58KK40 pKa = 10.44YY41 pKa = 9.73VQPQTKK47 pKa = 9.72NSQFFLSFLDD57 pKa = 3.38QCTMEE62 pKa = 4.51LAGIPGPSLEE72 pKa = 4.41ITTSEE77 pKa = 4.12YY78 pKa = 11.04VPPSEE83 pKa = 4.67SDD85 pKa = 3.41LFAHH89 pKa = 6.94IAGFGEE95 pKa = 4.6SGSSGSQLSNRR106 pKa = 11.84VYY108 pKa = 10.25AALRR112 pKa = 11.84EE113 pKa = 4.12EE114 pKa = 4.44LTMIEE119 pKa = 3.83HH120 pKa = 7.73WDD122 pKa = 3.72RR123 pKa = 11.84EE124 pKa = 4.25ITGWLDD130 pKa = 2.96PRR132 pKa = 11.84LLVDD136 pKa = 4.32IKK138 pKa = 11.23VPSQTSPGIRR148 pKa = 11.84WKK150 pKa = 10.87KK151 pKa = 10.3LGYY154 pKa = 8.14KK155 pKa = 7.97TKK157 pKa = 10.78KK158 pKa = 8.32EE159 pKa = 4.08ALMPATMEE167 pKa = 4.03AAKK170 pKa = 10.57VLLSMIEE177 pKa = 3.88TGTEE181 pKa = 4.23YY182 pKa = 10.41EE183 pKa = 4.34VPPCGVAGRR192 pKa = 11.84GKK194 pKa = 10.61RR195 pKa = 11.84MDD197 pKa = 3.56MNRR200 pKa = 11.84KK201 pKa = 9.27VDD203 pKa = 3.44TGGKK207 pKa = 9.41KK208 pKa = 9.3EE209 pKa = 3.84GRR211 pKa = 11.84LIVMPDD217 pKa = 3.54LVRR220 pKa = 11.84HH221 pKa = 5.67LVGTLGSGAYY231 pKa = 7.84MSKK234 pKa = 10.36VKK236 pKa = 10.44EE237 pKa = 3.97VDD239 pKa = 3.04KK240 pKa = 11.27SQGGILLGMGPFSEE254 pKa = 5.18SYY256 pKa = 10.74QSIANWARR264 pKa = 11.84GASKK268 pKa = 10.2FVFLDD273 pKa = 4.22FKK275 pKa = 11.23KK276 pKa = 10.43FDD278 pKa = 3.43HH279 pKa = 6.42RR280 pKa = 11.84VPARR284 pKa = 11.84VLQEE288 pKa = 4.04VFKK291 pKa = 10.68HH292 pKa = 4.7IASRR296 pKa = 11.84YY297 pKa = 7.05EE298 pKa = 3.78GCPGSKK304 pKa = 10.13AYY306 pKa = 8.89WRR308 pKa = 11.84SEE310 pKa = 3.56FQQLVNTKK318 pKa = 9.77IAMPDD323 pKa = 3.03GTVYY327 pKa = 10.06QKK329 pKa = 10.96KK330 pKa = 10.09RR331 pKa = 11.84GVASGDD337 pKa = 3.07PWTSIAGSYY346 pKa = 9.19ANWMMLKK353 pKa = 10.06HH354 pKa = 5.46VCNVLGLVAKK364 pKa = 10.15IWTFGDD370 pKa = 3.47DD371 pKa = 4.1SVIALYY377 pKa = 10.7DD378 pKa = 3.46VDD380 pKa = 4.23PTLDD384 pKa = 4.07LLPLFTKK391 pKa = 10.52HH392 pKa = 5.95LASSFGMHH400 pKa = 6.18VSQEE404 pKa = 3.66KK405 pKa = 10.75SYY407 pKa = 11.71VSDD410 pKa = 3.85CLVDD414 pKa = 3.28IAEE417 pKa = 4.55DD418 pKa = 4.14PEE420 pKa = 4.63PKK422 pKa = 10.25SSGSFLSLYY431 pKa = 10.3FLQTPMGVRR440 pKa = 11.84PTRR443 pKa = 11.84PLQDD447 pKa = 3.56MYY449 pKa = 11.6EE450 pKa = 3.96MMMVPEE456 pKa = 4.77RR457 pKa = 11.84NQGTLRR463 pKa = 11.84WEE465 pKa = 4.09IVRR468 pKa = 11.84TSMAYY473 pKa = 8.65LTFYY477 pKa = 11.04YY478 pKa = 10.23NDD480 pKa = 3.45RR481 pKa = 11.84ARR483 pKa = 11.84YY484 pKa = 8.91VLEE487 pKa = 5.15EE488 pKa = 3.97YY489 pKa = 10.1WDD491 pKa = 3.58WLHH494 pKa = 6.83RR495 pKa = 11.84KK496 pKa = 9.01YY497 pKa = 10.39RR498 pKa = 11.84VPEE501 pKa = 3.87LKK503 pKa = 11.06GNFTDD508 pKa = 5.06LNLLRR513 pKa = 11.84EE514 pKa = 4.05MDD516 pKa = 4.02IPWSSFKK523 pKa = 11.3LEE525 pKa = 3.88WLVRR529 pKa = 11.84LPRR532 pKa = 11.84PGEE535 pKa = 3.85VEE537 pKa = 4.5LMYY540 pKa = 10.61KK541 pKa = 10.19YY542 pKa = 10.78GHH544 pKa = 6.54TGFFPPLLWGHH555 pKa = 6.64WYY557 pKa = 10.37ARR559 pKa = 11.84YY560 pKa = 10.17DD561 pKa = 3.89RR562 pKa = 11.84DD563 pKa = 3.56VHH565 pKa = 6.22GNNVSFAFQEE575 pKa = 4.46PPTT578 pKa = 4.09
Molecular weight: 65.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
902 |
324 |
578 |
451.0 |
50.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.647 ± 3.47 | 0.776 ± 0.277 |
4.767 ± 0.264 | 6.984 ± 0.616 |
3.991 ± 0.902 | 7.871 ± 0.275 |
1.441 ± 0.671 | 3.215 ± 0.076 |
5.322 ± 1.324 | 9.867 ± 0.006 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.772 ± 1.276 | 2.106 ± 0.151 |
6.984 ± 1.165 | 3.437 ± 0.524 |
6.763 ± 1.296 | 6.652 ± 1.198 |
5.1 ± 0.27 | 7.539 ± 0.653 |
2.217 ± 0.582 | 3.548 ± 1.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
