
Altericroceibacterium endophyticum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Altericroceibacterium
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
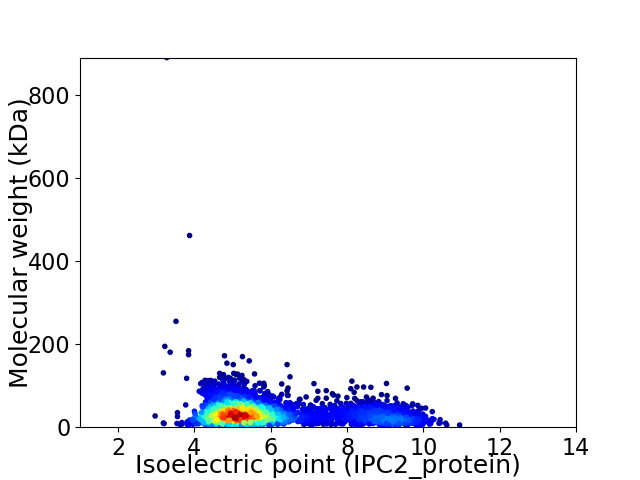
Virtual 2D-PAGE plot for 3079 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I4T3V5|A0A6I4T3V5_9SPHN Type I secretion system permease/ATPase OS=Altericroceibacterium endophyticum OX=1808508 GN=GRI91_09325 PE=4 SV=1
MM1 pKa = 7.36FMATANIAHH10 pKa = 7.45AGTCTEE16 pKa = 4.19TAPGSGDD23 pKa = 3.4WTCSGAANSGSDD35 pKa = 3.47TPEE38 pKa = 3.67TLGNGTGSEE47 pKa = 4.64LIVTTASGFGIDD59 pKa = 4.09NASGTAFILTNTAGGNNLTFTDD81 pKa = 5.21AYY83 pKa = 10.61ASDD86 pKa = 3.45ISGTGNGIFAVNNGTGITSITTTGVVTGTGTGTYY120 pKa = 10.27GIYY123 pKa = 10.56ARR125 pKa = 11.84NLGTDD130 pKa = 3.85LTISAADD137 pKa = 3.35VSGADD142 pKa = 4.22GISVANLGTGSTSVTATGTVTGTIYY167 pKa = 10.62RR168 pKa = 11.84GIVVNNVSTSTNMTVSAVDD187 pKa = 3.24VSGGTDD193 pKa = 4.28GIYY196 pKa = 10.75AFNNGTGFNSVTATGTVTGSAGSGILAVNYY226 pKa = 7.54GTNLTVSAVDD236 pKa = 3.49ASGSNSGIDD245 pKa = 3.32ALNYY249 pKa = 7.69GTGSNSVTATGTVTGNTNYY268 pKa = 10.78GINARR273 pKa = 11.84NFANSSTDD281 pKa = 3.36LTVFAADD288 pKa = 3.17ASGPNVGIFAYY299 pKa = 10.91NNGTGSTDD307 pKa = 3.05VTATGTVTSTINSGINASNSSNATNLTVSAVDD339 pKa = 3.3VSGGTDD345 pKa = 4.44GIRR348 pKa = 11.84AINNGSGSNSITATGTVTGTINYY371 pKa = 7.95GINVFGNSTATNLTVSAANASGGTNGIVTYY401 pKa = 10.98NSGTGSTRR409 pKa = 11.84VTATGTVAGTTSYY422 pKa = 11.03GISAWNDD429 pKa = 2.96STATDD434 pKa = 4.07LTVSAADD441 pKa = 3.34VSGGYY446 pKa = 9.63IGIEE450 pKa = 3.97AYY452 pKa = 11.0NNGTGTTSVTTTGAVSAVNAGILVYY477 pKa = 10.63SQNDD481 pKa = 3.61TFAVSVDD488 pKa = 3.74SGSVAAVGGGNDD500 pKa = 3.1AYY502 pKa = 10.83ARR504 pKa = 11.84AIQTSGAKK512 pKa = 10.27GGTIDD517 pKa = 3.27IAAGAVVDD525 pKa = 5.55GSASGIAIRR534 pKa = 11.84DD535 pKa = 3.15GDD537 pKa = 4.18YY538 pKa = 10.68ATGLGDD544 pKa = 4.08VLDD547 pKa = 4.56GTDD550 pKa = 3.49LTGGDD555 pKa = 4.0VVVTTYY561 pKa = 11.78GDD563 pKa = 3.56VKK565 pKa = 11.08GDD567 pKa = 4.13AILGKK572 pKa = 11.07GNDD575 pKa = 3.75TFNLAGGDD583 pKa = 3.92FDD585 pKa = 5.64GDD587 pKa = 3.43IYY589 pKa = 11.36GDD591 pKa = 3.64DD592 pKa = 3.66TLASVNDD599 pKa = 4.08GNDD602 pKa = 3.07TFNFTGGDD610 pKa = 3.03WHH612 pKa = 8.18SGFFGGNGSDD622 pKa = 3.47TANISSPTYY631 pKa = 10.54DD632 pKa = 3.13GTAHH636 pKa = 6.31VLDD639 pKa = 5.13GGDD642 pKa = 4.49DD643 pKa = 3.75YY644 pKa = 11.75GTGDD648 pKa = 3.25GWIDD652 pKa = 3.42TLTFAGVTSDD662 pKa = 4.67ANGTNILNWEE672 pKa = 4.32NIVINGGAITIVDD685 pKa = 3.94GALEE689 pKa = 4.31TGSDD693 pKa = 3.44AGTGLTVMNGGVLDD707 pKa = 4.56GSDD710 pKa = 3.52ALALTGNLVIKK721 pKa = 10.71SGGAFVGTGGGAGVYY736 pKa = 10.02SISGDD741 pKa = 3.29VSNQGTITTQDD752 pKa = 3.19GVVGDD757 pKa = 3.98VVSIGGNYY765 pKa = 9.87SGGGTITVDD774 pKa = 3.67ANLDD778 pKa = 3.68GAGSADD784 pKa = 3.51QIVIAGNVTGPATTVNLNNVGSGIISPTTGNGIFVVSAGGTSSAGSFVLNPSDD837 pKa = 3.7AQVGAFDD844 pKa = 3.92YY845 pKa = 11.08SINQASDD852 pKa = 2.87GKK854 pKa = 10.8LYY856 pKa = 10.97LSSTYY861 pKa = 9.82QAGVPVMEE869 pKa = 5.86AYY871 pKa = 9.94GQALLGLNSLNSLRR885 pKa = 11.84QRR887 pKa = 11.84TGNRR891 pKa = 11.84SWTPGLSKK899 pKa = 9.91TIDD902 pKa = 3.74DD903 pKa = 4.02TSTAAGVWGQFEE915 pKa = 4.41GSLAEE920 pKa = 4.12QNFATSVTGSAYY932 pKa = 10.5DD933 pKa = 3.32QDD935 pKa = 3.81FWRR938 pKa = 11.84LRR940 pKa = 11.84TGVEE944 pKa = 4.02APLTLSDD951 pKa = 3.9SGALFVGTQVNYY963 pKa = 10.29GVARR967 pKa = 11.84TNVTSVYY974 pKa = 10.06GNGRR978 pKa = 11.84IMTNVLGFGLGMTWYY993 pKa = 10.07GNSGFYY999 pKa = 10.95ADD1001 pKa = 3.05IQGRR1005 pKa = 11.84YY1006 pKa = 7.02NHH1008 pKa = 6.3YY1009 pKa = 10.86SSDD1012 pKa = 3.22VRR1014 pKa = 11.84NGLGRR1019 pKa = 11.84MANNVKK1025 pKa = 10.39SDD1027 pKa = 3.33GYY1029 pKa = 11.44AGGLEE1034 pKa = 4.18LGYY1037 pKa = 11.16VLAQGNSWKK1046 pKa = 8.68ITPQAQLTYY1055 pKa = 9.62GTVDD1059 pKa = 3.29YY1060 pKa = 11.53NSFSDD1065 pKa = 4.02GTSATAALADD1075 pKa = 3.69GSVNSWRR1082 pKa = 11.84GRR1084 pKa = 11.84GGLEE1088 pKa = 4.92LSNDD1092 pKa = 3.86KK1093 pKa = 10.91NWTSEE1098 pKa = 3.81SGKK1101 pKa = 10.12EE1102 pKa = 4.07MSRR1105 pKa = 11.84HH1106 pKa = 5.16FYY1108 pKa = 10.85GVLNLHH1114 pKa = 6.18YY1115 pKa = 10.53EE1116 pKa = 4.24FDD1118 pKa = 4.17GKK1120 pKa = 11.41SNAAVSNTPLYY1131 pKa = 10.64SRR1133 pKa = 11.84PEE1135 pKa = 4.02QLWGEE1140 pKa = 4.3VGAGFQHH1147 pKa = 6.27NAVGSISVFGEE1158 pKa = 3.52AAYY1161 pKa = 10.53SVSFANGGDD1170 pKa = 3.69NNKK1173 pKa = 10.24LNGRR1177 pKa = 11.84IGLRR1181 pKa = 11.84ANFF1184 pKa = 3.78
MM1 pKa = 7.36FMATANIAHH10 pKa = 7.45AGTCTEE16 pKa = 4.19TAPGSGDD23 pKa = 3.4WTCSGAANSGSDD35 pKa = 3.47TPEE38 pKa = 3.67TLGNGTGSEE47 pKa = 4.64LIVTTASGFGIDD59 pKa = 4.09NASGTAFILTNTAGGNNLTFTDD81 pKa = 5.21AYY83 pKa = 10.61ASDD86 pKa = 3.45ISGTGNGIFAVNNGTGITSITTTGVVTGTGTGTYY120 pKa = 10.27GIYY123 pKa = 10.56ARR125 pKa = 11.84NLGTDD130 pKa = 3.85LTISAADD137 pKa = 3.35VSGADD142 pKa = 4.22GISVANLGTGSTSVTATGTVTGTIYY167 pKa = 10.62RR168 pKa = 11.84GIVVNNVSTSTNMTVSAVDD187 pKa = 3.24VSGGTDD193 pKa = 4.28GIYY196 pKa = 10.75AFNNGTGFNSVTATGTVTGSAGSGILAVNYY226 pKa = 7.54GTNLTVSAVDD236 pKa = 3.49ASGSNSGIDD245 pKa = 3.32ALNYY249 pKa = 7.69GTGSNSVTATGTVTGNTNYY268 pKa = 10.78GINARR273 pKa = 11.84NFANSSTDD281 pKa = 3.36LTVFAADD288 pKa = 3.17ASGPNVGIFAYY299 pKa = 10.91NNGTGSTDD307 pKa = 3.05VTATGTVTSTINSGINASNSSNATNLTVSAVDD339 pKa = 3.3VSGGTDD345 pKa = 4.44GIRR348 pKa = 11.84AINNGSGSNSITATGTVTGTINYY371 pKa = 7.95GINVFGNSTATNLTVSAANASGGTNGIVTYY401 pKa = 10.98NSGTGSTRR409 pKa = 11.84VTATGTVAGTTSYY422 pKa = 11.03GISAWNDD429 pKa = 2.96STATDD434 pKa = 4.07LTVSAADD441 pKa = 3.34VSGGYY446 pKa = 9.63IGIEE450 pKa = 3.97AYY452 pKa = 11.0NNGTGTTSVTTTGAVSAVNAGILVYY477 pKa = 10.63SQNDD481 pKa = 3.61TFAVSVDD488 pKa = 3.74SGSVAAVGGGNDD500 pKa = 3.1AYY502 pKa = 10.83ARR504 pKa = 11.84AIQTSGAKK512 pKa = 10.27GGTIDD517 pKa = 3.27IAAGAVVDD525 pKa = 5.55GSASGIAIRR534 pKa = 11.84DD535 pKa = 3.15GDD537 pKa = 4.18YY538 pKa = 10.68ATGLGDD544 pKa = 4.08VLDD547 pKa = 4.56GTDD550 pKa = 3.49LTGGDD555 pKa = 4.0VVVTTYY561 pKa = 11.78GDD563 pKa = 3.56VKK565 pKa = 11.08GDD567 pKa = 4.13AILGKK572 pKa = 11.07GNDD575 pKa = 3.75TFNLAGGDD583 pKa = 3.92FDD585 pKa = 5.64GDD587 pKa = 3.43IYY589 pKa = 11.36GDD591 pKa = 3.64DD592 pKa = 3.66TLASVNDD599 pKa = 4.08GNDD602 pKa = 3.07TFNFTGGDD610 pKa = 3.03WHH612 pKa = 8.18SGFFGGNGSDD622 pKa = 3.47TANISSPTYY631 pKa = 10.54DD632 pKa = 3.13GTAHH636 pKa = 6.31VLDD639 pKa = 5.13GGDD642 pKa = 4.49DD643 pKa = 3.75YY644 pKa = 11.75GTGDD648 pKa = 3.25GWIDD652 pKa = 3.42TLTFAGVTSDD662 pKa = 4.67ANGTNILNWEE672 pKa = 4.32NIVINGGAITIVDD685 pKa = 3.94GALEE689 pKa = 4.31TGSDD693 pKa = 3.44AGTGLTVMNGGVLDD707 pKa = 4.56GSDD710 pKa = 3.52ALALTGNLVIKK721 pKa = 10.71SGGAFVGTGGGAGVYY736 pKa = 10.02SISGDD741 pKa = 3.29VSNQGTITTQDD752 pKa = 3.19GVVGDD757 pKa = 3.98VVSIGGNYY765 pKa = 9.87SGGGTITVDD774 pKa = 3.67ANLDD778 pKa = 3.68GAGSADD784 pKa = 3.51QIVIAGNVTGPATTVNLNNVGSGIISPTTGNGIFVVSAGGTSSAGSFVLNPSDD837 pKa = 3.7AQVGAFDD844 pKa = 3.92YY845 pKa = 11.08SINQASDD852 pKa = 2.87GKK854 pKa = 10.8LYY856 pKa = 10.97LSSTYY861 pKa = 9.82QAGVPVMEE869 pKa = 5.86AYY871 pKa = 9.94GQALLGLNSLNSLRR885 pKa = 11.84QRR887 pKa = 11.84TGNRR891 pKa = 11.84SWTPGLSKK899 pKa = 9.91TIDD902 pKa = 3.74DD903 pKa = 4.02TSTAAGVWGQFEE915 pKa = 4.41GSLAEE920 pKa = 4.12QNFATSVTGSAYY932 pKa = 10.5DD933 pKa = 3.32QDD935 pKa = 3.81FWRR938 pKa = 11.84LRR940 pKa = 11.84TGVEE944 pKa = 4.02APLTLSDD951 pKa = 3.9SGALFVGTQVNYY963 pKa = 10.29GVARR967 pKa = 11.84TNVTSVYY974 pKa = 10.06GNGRR978 pKa = 11.84IMTNVLGFGLGMTWYY993 pKa = 10.07GNSGFYY999 pKa = 10.95ADD1001 pKa = 3.05IQGRR1005 pKa = 11.84YY1006 pKa = 7.02NHH1008 pKa = 6.3YY1009 pKa = 10.86SSDD1012 pKa = 3.22VRR1014 pKa = 11.84NGLGRR1019 pKa = 11.84MANNVKK1025 pKa = 10.39SDD1027 pKa = 3.33GYY1029 pKa = 11.44AGGLEE1034 pKa = 4.18LGYY1037 pKa = 11.16VLAQGNSWKK1046 pKa = 8.68ITPQAQLTYY1055 pKa = 9.62GTVDD1059 pKa = 3.29YY1060 pKa = 11.53NSFSDD1065 pKa = 4.02GTSATAALADD1075 pKa = 3.69GSVNSWRR1082 pKa = 11.84GRR1084 pKa = 11.84GGLEE1088 pKa = 4.92LSNDD1092 pKa = 3.86KK1093 pKa = 10.91NWTSEE1098 pKa = 3.81SGKK1101 pKa = 10.12EE1102 pKa = 4.07MSRR1105 pKa = 11.84HH1106 pKa = 5.16FYY1108 pKa = 10.85GVLNLHH1114 pKa = 6.18YY1115 pKa = 10.53EE1116 pKa = 4.24FDD1118 pKa = 4.17GKK1120 pKa = 11.41SNAAVSNTPLYY1131 pKa = 10.64SRR1133 pKa = 11.84PEE1135 pKa = 4.02QLWGEE1140 pKa = 4.3VGAGFQHH1147 pKa = 6.27NAVGSISVFGEE1158 pKa = 3.52AAYY1161 pKa = 10.53SVSFANGGDD1170 pKa = 3.69NNKK1173 pKa = 10.24LNGRR1177 pKa = 11.84IGLRR1181 pKa = 11.84ANFF1184 pKa = 3.78
Molecular weight: 117.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I4T5C2|A0A6I4T5C2_9SPHN Helix-turn-helix domain-containing protein OS=Altericroceibacterium endophyticum OX=1808508 GN=GRI91_06520 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.51GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.83ATVGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.97RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.46NLCAA44 pKa = 4.54
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.51GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.83ATVGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.97RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.46NLCAA44 pKa = 4.54
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1041826 |
36 |
8661 |
338.4 |
36.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.886 ± 0.062 | 0.817 ± 0.014 |
6.356 ± 0.07 | 5.966 ± 0.041 |
3.713 ± 0.032 | 8.739 ± 0.097 |
1.941 ± 0.028 | 5.278 ± 0.031 |
3.22 ± 0.039 | 9.775 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.524 ± 0.028 | 2.983 ± 0.034 |
5.059 ± 0.039 | 3.515 ± 0.024 |
6.61 ± 0.053 | 5.939 ± 0.037 |
5.225 ± 0.036 | 6.605 ± 0.04 |
1.425 ± 0.025 | 2.424 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
