
Rift valley fever virus (strain ZH-548 M12) (RVFV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Rift Valley fever phlebovirus
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
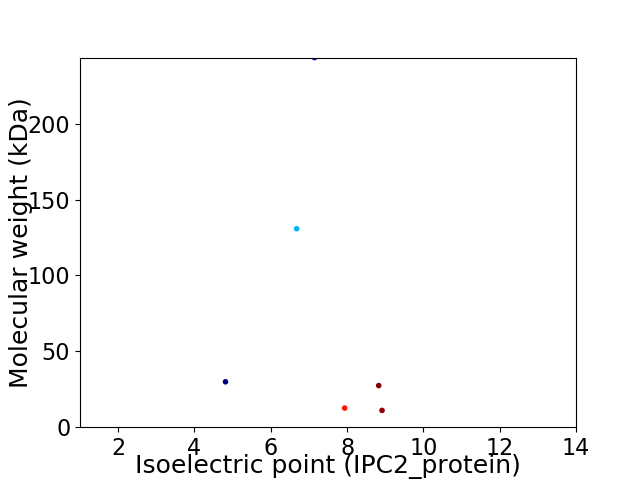
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P21700|NCAP_RVFVZ Nucleoprotein OS=Rift valley fever virus (strain ZH-548 M12) OX=11589 GN=N PE=1 SV=1
MM1 pKa = 8.36DD2 pKa = 4.9YY3 pKa = 10.96FPVISVDD10 pKa = 3.68LQSGRR15 pKa = 11.84RR16 pKa = 11.84VVSVEE21 pKa = 3.54YY22 pKa = 10.58FRR24 pKa = 11.84GDD26 pKa = 3.39GPPRR30 pKa = 11.84IPYY33 pKa = 10.8SMVGPCCVFLMHH45 pKa = 6.78HH46 pKa = 6.51RR47 pKa = 11.84PSHH50 pKa = 4.79EE51 pKa = 3.6VRR53 pKa = 11.84LRR55 pKa = 11.84FSDD58 pKa = 4.43FYY60 pKa = 11.69NVGEE64 pKa = 3.95FPYY67 pKa = 10.48RR68 pKa = 11.84VGLGDD73 pKa = 3.64FASNVAPPPAKK84 pKa = 10.09PFQRR88 pKa = 11.84LIDD91 pKa = 4.95LIGHH95 pKa = 5.42MTLSDD100 pKa = 3.52FTRR103 pKa = 11.84FPNLKK108 pKa = 9.9EE109 pKa = 4.39AISWPLGEE117 pKa = 4.82PSLAFFDD124 pKa = 4.54LSSTRR129 pKa = 11.84VHH131 pKa = 6.71RR132 pKa = 11.84NDD134 pKa = 5.09DD135 pKa = 3.07IRR137 pKa = 11.84RR138 pKa = 11.84DD139 pKa = 3.87QIATLAMRR147 pKa = 11.84SCKK150 pKa = 8.73ITNDD154 pKa = 3.67LEE156 pKa = 4.43DD157 pKa = 4.02SFVGLHH163 pKa = 6.38RR164 pKa = 11.84MIATEE169 pKa = 4.63AILRR173 pKa = 11.84GIDD176 pKa = 3.45LCLLPGFDD184 pKa = 4.68LMYY187 pKa = 10.65EE188 pKa = 4.3VAHH191 pKa = 5.14VQCVRR196 pKa = 11.84LLQAAKK202 pKa = 10.31EE203 pKa = 4.41DD204 pKa = 3.32ISNAVVPNSALIVLMEE220 pKa = 4.43EE221 pKa = 4.29SLMLRR226 pKa = 11.84SSLPSMMGRR235 pKa = 11.84NNWIPVIPPIPDD247 pKa = 2.86VEE249 pKa = 4.31MEE251 pKa = 4.3SEE253 pKa = 4.35EE254 pKa = 4.48EE255 pKa = 3.98SDD257 pKa = 3.98DD258 pKa = 4.23DD259 pKa = 5.24GFVEE263 pKa = 4.86VDD265 pKa = 2.97
MM1 pKa = 8.36DD2 pKa = 4.9YY3 pKa = 10.96FPVISVDD10 pKa = 3.68LQSGRR15 pKa = 11.84RR16 pKa = 11.84VVSVEE21 pKa = 3.54YY22 pKa = 10.58FRR24 pKa = 11.84GDD26 pKa = 3.39GPPRR30 pKa = 11.84IPYY33 pKa = 10.8SMVGPCCVFLMHH45 pKa = 6.78HH46 pKa = 6.51RR47 pKa = 11.84PSHH50 pKa = 4.79EE51 pKa = 3.6VRR53 pKa = 11.84LRR55 pKa = 11.84FSDD58 pKa = 4.43FYY60 pKa = 11.69NVGEE64 pKa = 3.95FPYY67 pKa = 10.48RR68 pKa = 11.84VGLGDD73 pKa = 3.64FASNVAPPPAKK84 pKa = 10.09PFQRR88 pKa = 11.84LIDD91 pKa = 4.95LIGHH95 pKa = 5.42MTLSDD100 pKa = 3.52FTRR103 pKa = 11.84FPNLKK108 pKa = 9.9EE109 pKa = 4.39AISWPLGEE117 pKa = 4.82PSLAFFDD124 pKa = 4.54LSSTRR129 pKa = 11.84VHH131 pKa = 6.71RR132 pKa = 11.84NDD134 pKa = 5.09DD135 pKa = 3.07IRR137 pKa = 11.84RR138 pKa = 11.84DD139 pKa = 3.87QIATLAMRR147 pKa = 11.84SCKK150 pKa = 8.73ITNDD154 pKa = 3.67LEE156 pKa = 4.43DD157 pKa = 4.02SFVGLHH163 pKa = 6.38RR164 pKa = 11.84MIATEE169 pKa = 4.63AILRR173 pKa = 11.84GIDD176 pKa = 3.45LCLLPGFDD184 pKa = 4.68LMYY187 pKa = 10.65EE188 pKa = 4.3VAHH191 pKa = 5.14VQCVRR196 pKa = 11.84LLQAAKK202 pKa = 10.31EE203 pKa = 4.41DD204 pKa = 3.32ISNAVVPNSALIVLMEE220 pKa = 4.43EE221 pKa = 4.29SLMLRR226 pKa = 11.84SSLPSMMGRR235 pKa = 11.84NNWIPVIPPIPDD247 pKa = 2.86VEE249 pKa = 4.31MEE251 pKa = 4.3SEE253 pKa = 4.35EE254 pKa = 4.48EE255 pKa = 3.98SDD257 pKa = 3.98DD258 pKa = 4.23DD259 pKa = 5.24GFVEE263 pKa = 4.86VDD265 pKa = 2.97
Molecular weight: 29.93 kDa
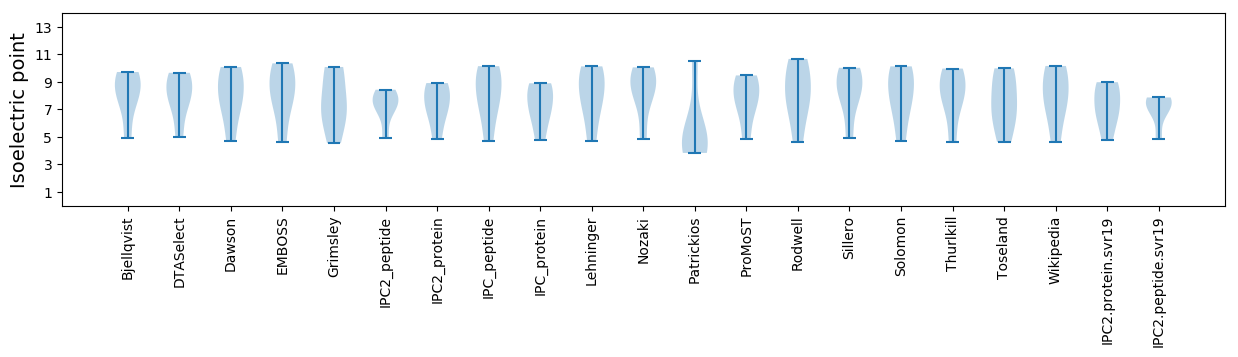
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P21401-5|GP-5_RVFVZ Isoform of P21401 Isoform NSm' protein of Envelopment polyprotein OS=Rift valley fever virus (strain ZH-548 M12) OX=11589 GN=GP PE=1 SV=1
MM1 pKa = 7.79PEE3 pKa = 4.33EE4 pKa = 4.46LSCSISGIRR13 pKa = 11.84EE14 pKa = 4.08VKK16 pKa = 9.61TSSQEE21 pKa = 3.56LYY23 pKa = 10.04RR24 pKa = 11.84ALKK27 pKa = 10.43AIIAADD33 pKa = 3.58GLNNITCHH41 pKa = 6.42GKK43 pKa = 10.26DD44 pKa = 3.6PEE46 pKa = 4.67DD47 pKa = 4.53KK48 pKa = 10.41ISLIKK53 pKa = 10.62GPPHH57 pKa = 6.89KK58 pKa = 10.26KK59 pKa = 9.01RR60 pKa = 11.84VGIVRR65 pKa = 11.84CEE67 pKa = 3.61RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84DD71 pKa = 3.3AKK73 pKa = 10.8QIGRR77 pKa = 11.84KK78 pKa = 7.15TMAGIAMTVLPALAVFALAPVVFAA102 pKa = 5.8
MM1 pKa = 7.79PEE3 pKa = 4.33EE4 pKa = 4.46LSCSISGIRR13 pKa = 11.84EE14 pKa = 4.08VKK16 pKa = 9.61TSSQEE21 pKa = 3.56LYY23 pKa = 10.04RR24 pKa = 11.84ALKK27 pKa = 10.43AIIAADD33 pKa = 3.58GLNNITCHH41 pKa = 6.42GKK43 pKa = 10.26DD44 pKa = 3.6PEE46 pKa = 4.67DD47 pKa = 4.53KK48 pKa = 10.41ISLIKK53 pKa = 10.62GPPHH57 pKa = 6.89KK58 pKa = 10.26KK59 pKa = 9.01RR60 pKa = 11.84VGIVRR65 pKa = 11.84CEE67 pKa = 3.61RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84DD71 pKa = 3.3AKK73 pKa = 10.8QIGRR77 pKa = 11.84KK78 pKa = 7.15TMAGIAMTVLPALAVFALAPVVFAA102 pKa = 5.8
Molecular weight: 11.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4073 |
102 |
2149 |
678.8 |
75.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.85 ± 1.237 | 2.946 ± 0.868 |
5.009 ± 0.404 | 6.359 ± 0.326 |
4.591 ± 0.493 | 6.31 ± 0.554 |
2.431 ± 0.15 | 6.187 ± 0.565 |
6.113 ± 0.589 | 9.305 ± 0.292 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.192 ± 0.533 | 3.585 ± 0.246 |
4.493 ± 0.588 | 3.094 ± 0.281 |
5.573 ± 0.773 | 8.618 ± 0.738 |
4.739 ± 0.519 | 6.752 ± 0.297 |
1.154 ± 0.13 | 2.701 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
