
Acinetobacter calcoaceticus (strain PHEA-2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Acinetobacter; Acinetobacter calcoaceticus/baumannii complex; Acinetobacter pittii
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
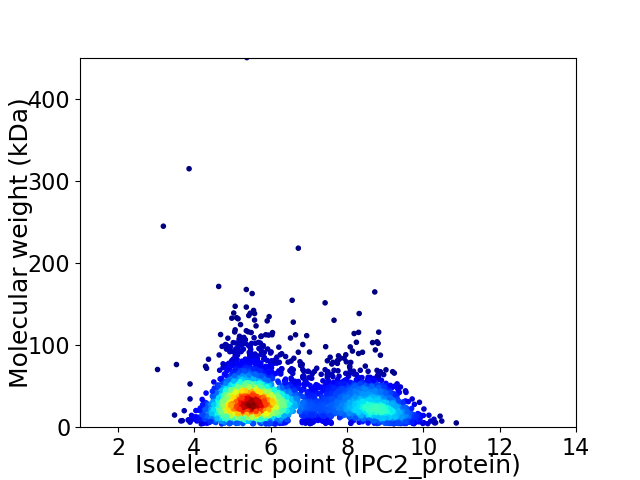
Virtual 2D-PAGE plot for 3598 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
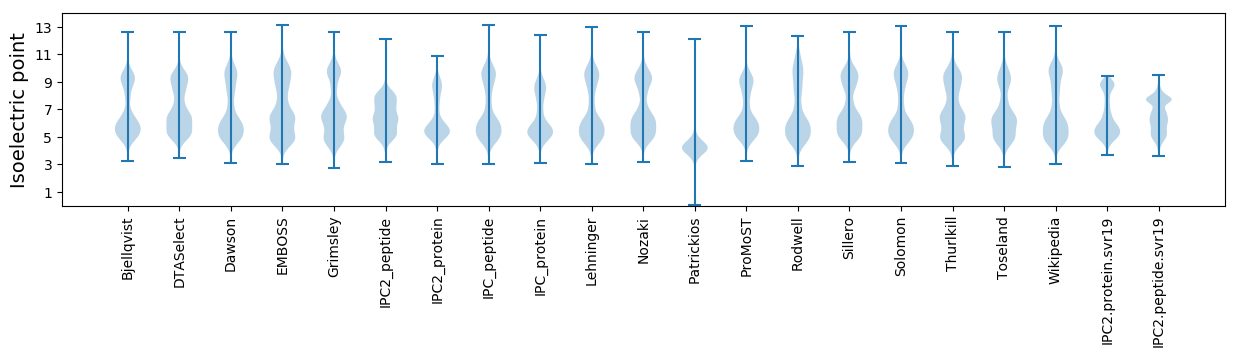
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0KPV5|F0KPV5_ACICP Uncharacterized protein OS=Acinetobacter calcoaceticus (strain PHEA-2) OX=871585 GN=BDGL_002464 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 9.67IDD4 pKa = 4.35NISYY8 pKa = 10.39EE9 pKa = 4.12YY10 pKa = 11.02DD11 pKa = 3.63DD12 pKa = 5.93LIKK15 pKa = 11.23LNITDD20 pKa = 3.87TKK22 pKa = 10.61EE23 pKa = 4.01IEE25 pKa = 5.06CMTLLNKK32 pKa = 9.88LDD34 pKa = 3.69AMTPILTNLFSYY46 pKa = 10.61DD47 pKa = 3.15VSTDD51 pKa = 3.52KK52 pKa = 10.96IIFNPPSDD60 pKa = 3.81SYY62 pKa = 11.3IVEE65 pKa = 4.12QKK67 pKa = 10.63VDD69 pKa = 3.74NTWVQINLEE78 pKa = 4.33EE79 pKa = 4.75KK80 pKa = 10.45FDD82 pKa = 3.78WLNTEE87 pKa = 4.28FRR89 pKa = 11.84VTTTDD94 pKa = 2.94QAGNISQPLITIINTASGTYY114 pKa = 9.93KK115 pKa = 9.83PTDD118 pKa = 3.34TTSNQMVKK126 pKa = 10.72GSTGNDD132 pKa = 3.24YY133 pKa = 11.18LYY135 pKa = 11.19GGDD138 pKa = 4.67GNDD141 pKa = 3.46TLISNTGSDD150 pKa = 3.97YY151 pKa = 11.18LYY153 pKa = 11.1GGAGNDD159 pKa = 3.44TLVYY163 pKa = 10.35GGNSNGYY170 pKa = 7.18TALLGQAGNDD180 pKa = 3.37TYY182 pKa = 11.03IIDD185 pKa = 3.99KK186 pKa = 10.73VLLSSLSYY194 pKa = 10.81VHH196 pKa = 7.18ILDD199 pKa = 4.2NSTEE203 pKa = 4.05EE204 pKa = 3.78NTLQLKK210 pKa = 9.99SVSADD215 pKa = 3.59EE216 pKa = 5.62IILKK220 pKa = 10.17QSNTDD225 pKa = 3.49IIITFNDD232 pKa = 3.18STATIHH238 pKa = 6.64FGEE241 pKa = 4.81GQLSAIVFDD250 pKa = 6.39DD251 pKa = 3.7GTVWNKK257 pKa = 10.2AQIEE261 pKa = 4.33ANTIGKK267 pKa = 9.36LLGNDD272 pKa = 3.3ADD274 pKa = 5.96DD275 pKa = 4.21YY276 pKa = 11.03LQADD280 pKa = 4.15AEE282 pKa = 4.2ISTIYY287 pKa = 10.52GLEE290 pKa = 4.28GNDD293 pKa = 3.79TIQGGVQNDD302 pKa = 3.67YY303 pKa = 11.38LYY305 pKa = 11.32GGDD308 pKa = 4.67GNDD311 pKa = 3.46TLISNTGTDD320 pKa = 3.72YY321 pKa = 11.44LYY323 pKa = 11.29GGAGNDD329 pKa = 3.43TLIYY333 pKa = 10.23GGNSNGYY340 pKa = 7.18TALLGQAGNDD350 pKa = 3.48TYY352 pKa = 11.16IVDD355 pKa = 4.0KK356 pKa = 10.98ALLSSWSYY364 pKa = 10.37VHH366 pKa = 7.25ILDD369 pKa = 4.76NSTEE373 pKa = 4.0EE374 pKa = 5.1NILQLKK380 pKa = 9.78SVLADD385 pKa = 4.01EE386 pKa = 5.82IILKK390 pKa = 10.3KK391 pKa = 10.85SNADD395 pKa = 3.44IIITFNDD402 pKa = 3.18STATIHH408 pKa = 6.84FGEE411 pKa = 4.52EE412 pKa = 3.9QLSSIVFDD420 pKa = 6.24DD421 pKa = 3.79GTVWNKK427 pKa = 10.2AQIEE431 pKa = 4.33ANTIGKK437 pKa = 9.33LLGTDD442 pKa = 3.22ADD444 pKa = 5.25DD445 pKa = 4.05YY446 pKa = 11.13LQADD450 pKa = 3.86AEE452 pKa = 4.47MKK454 pKa = 10.74NNYY457 pKa = 9.19LLLYY461 pKa = 9.62LTMEE465 pKa = 3.91QSGIKK470 pKa = 10.19LKK472 pKa = 10.82LKK474 pKa = 10.53RR475 pKa = 11.84ILLANYY481 pKa = 9.33
MM1 pKa = 7.56KK2 pKa = 9.67IDD4 pKa = 4.35NISYY8 pKa = 10.39EE9 pKa = 4.12YY10 pKa = 11.02DD11 pKa = 3.63DD12 pKa = 5.93LIKK15 pKa = 11.23LNITDD20 pKa = 3.87TKK22 pKa = 10.61EE23 pKa = 4.01IEE25 pKa = 5.06CMTLLNKK32 pKa = 9.88LDD34 pKa = 3.69AMTPILTNLFSYY46 pKa = 10.61DD47 pKa = 3.15VSTDD51 pKa = 3.52KK52 pKa = 10.96IIFNPPSDD60 pKa = 3.81SYY62 pKa = 11.3IVEE65 pKa = 4.12QKK67 pKa = 10.63VDD69 pKa = 3.74NTWVQINLEE78 pKa = 4.33EE79 pKa = 4.75KK80 pKa = 10.45FDD82 pKa = 3.78WLNTEE87 pKa = 4.28FRR89 pKa = 11.84VTTTDD94 pKa = 2.94QAGNISQPLITIINTASGTYY114 pKa = 9.93KK115 pKa = 9.83PTDD118 pKa = 3.34TTSNQMVKK126 pKa = 10.72GSTGNDD132 pKa = 3.24YY133 pKa = 11.18LYY135 pKa = 11.19GGDD138 pKa = 4.67GNDD141 pKa = 3.46TLISNTGSDD150 pKa = 3.97YY151 pKa = 11.18LYY153 pKa = 11.1GGAGNDD159 pKa = 3.44TLVYY163 pKa = 10.35GGNSNGYY170 pKa = 7.18TALLGQAGNDD180 pKa = 3.37TYY182 pKa = 11.03IIDD185 pKa = 3.99KK186 pKa = 10.73VLLSSLSYY194 pKa = 10.81VHH196 pKa = 7.18ILDD199 pKa = 4.2NSTEE203 pKa = 4.05EE204 pKa = 3.78NTLQLKK210 pKa = 9.99SVSADD215 pKa = 3.59EE216 pKa = 5.62IILKK220 pKa = 10.17QSNTDD225 pKa = 3.49IIITFNDD232 pKa = 3.18STATIHH238 pKa = 6.64FGEE241 pKa = 4.81GQLSAIVFDD250 pKa = 6.39DD251 pKa = 3.7GTVWNKK257 pKa = 10.2AQIEE261 pKa = 4.33ANTIGKK267 pKa = 9.36LLGNDD272 pKa = 3.3ADD274 pKa = 5.96DD275 pKa = 4.21YY276 pKa = 11.03LQADD280 pKa = 4.15AEE282 pKa = 4.2ISTIYY287 pKa = 10.52GLEE290 pKa = 4.28GNDD293 pKa = 3.79TIQGGVQNDD302 pKa = 3.67YY303 pKa = 11.38LYY305 pKa = 11.32GGDD308 pKa = 4.67GNDD311 pKa = 3.46TLISNTGTDD320 pKa = 3.72YY321 pKa = 11.44LYY323 pKa = 11.29GGAGNDD329 pKa = 3.43TLIYY333 pKa = 10.23GGNSNGYY340 pKa = 7.18TALLGQAGNDD350 pKa = 3.48TYY352 pKa = 11.16IVDD355 pKa = 4.0KK356 pKa = 10.98ALLSSWSYY364 pKa = 10.37VHH366 pKa = 7.25ILDD369 pKa = 4.76NSTEE373 pKa = 4.0EE374 pKa = 5.1NILQLKK380 pKa = 9.78SVLADD385 pKa = 4.01EE386 pKa = 5.82IILKK390 pKa = 10.3KK391 pKa = 10.85SNADD395 pKa = 3.44IIITFNDD402 pKa = 3.18STATIHH408 pKa = 6.84FGEE411 pKa = 4.52EE412 pKa = 3.9QLSSIVFDD420 pKa = 6.24DD421 pKa = 3.79GTVWNKK427 pKa = 10.2AQIEE431 pKa = 4.33ANTIGKK437 pKa = 9.33LLGTDD442 pKa = 3.22ADD444 pKa = 5.25DD445 pKa = 4.05YY446 pKa = 11.13LQADD450 pKa = 3.86AEE452 pKa = 4.47MKK454 pKa = 10.74NNYY457 pKa = 9.19LLLYY461 pKa = 9.62LTMEE465 pKa = 3.91QSGIKK470 pKa = 10.19LKK472 pKa = 10.82LKK474 pKa = 10.53RR475 pKa = 11.84ILLANYY481 pKa = 9.33
Molecular weight: 52.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0KPV4|F0KPV4_ACICP ClpXP protease specificity-enhancing factor OS=Acinetobacter calcoaceticus (strain PHEA-2) OX=871585 GN=sspB PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.55RR3 pKa = 11.84TFQPSEE9 pKa = 3.97LKK11 pKa = 10.13RR12 pKa = 11.84KK13 pKa = 8.98RR14 pKa = 11.84VHH16 pKa = 6.36GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4AGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.1SLTVV44 pKa = 3.06
MM1 pKa = 7.45KK2 pKa = 9.55RR3 pKa = 11.84TFQPSEE9 pKa = 3.97LKK11 pKa = 10.13RR12 pKa = 11.84KK13 pKa = 8.98RR14 pKa = 11.84VHH16 pKa = 6.36GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4AGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.1SLTVV44 pKa = 3.06
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1122070 |
37 |
4331 |
311.9 |
34.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.608 ± 0.049 | 0.934 ± 0.013 |
5.066 ± 0.033 | 5.862 ± 0.047 |
4.34 ± 0.034 | 6.713 ± 0.047 |
2.345 ± 0.023 | 6.855 ± 0.04 |
5.609 ± 0.037 | 10.473 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.025 | 4.485 ± 0.044 |
4.021 ± 0.029 | 5.268 ± 0.048 |
4.191 ± 0.037 | 6.141 ± 0.031 |
5.41 ± 0.042 | 6.733 ± 0.042 |
1.279 ± 0.016 | 3.27 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
