
Hamster polyomavirus (HaPyV) (Mesocricetus auratus polyomavirus 1)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
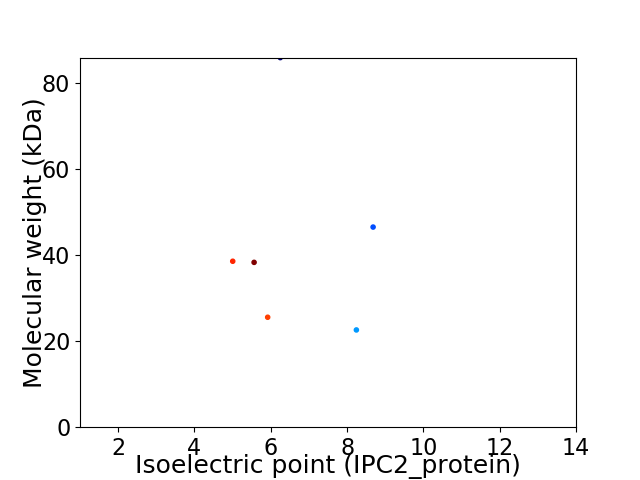
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
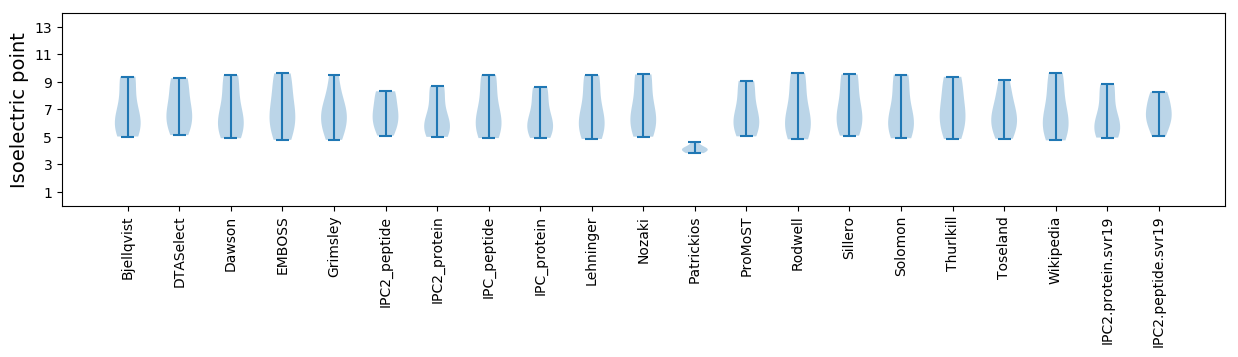
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q84255|Q84255_POVHA Capsid protein VP1 OS=Hamster polyomavirus OX=1891729 GN=VP1 PE=3 SV=1
MM1 pKa = 7.51GSAISVIIEE10 pKa = 4.14MISYY14 pKa = 10.09LSEE17 pKa = 3.78ISSVTGISVEE27 pKa = 4.46AILSGEE33 pKa = 3.91AFAAIDD39 pKa = 3.7AQVTSLITMEE49 pKa = 4.51GFLGAEE55 pKa = 4.29TALSSIGLSEE65 pKa = 4.76DD66 pKa = 3.07MFIFMQAAPEE76 pKa = 4.07LTSTVMTEE84 pKa = 4.26FVRR87 pKa = 11.84EE88 pKa = 4.16SVQTAFIFQTVAGSAAFSLGSLHH111 pKa = 7.19GYY113 pKa = 8.7LAHH116 pKa = 6.8EE117 pKa = 4.35VPIVNRR123 pKa = 11.84NMALIPRR130 pKa = 11.84RR131 pKa = 11.84PADD134 pKa = 3.37YY135 pKa = 11.21YY136 pKa = 11.28DD137 pKa = 3.24ILFPGVQSFTHH148 pKa = 6.32ALDD151 pKa = 4.59VIHH154 pKa = 6.89GWGHH158 pKa = 6.21SLFQSVGEE166 pKa = 4.51YY167 pKa = 9.89IWDD170 pKa = 3.59TLRR173 pKa = 11.84RR174 pKa = 11.84EE175 pKa = 4.04TQGAVEE181 pKa = 4.18SAVRR185 pKa = 11.84DD186 pKa = 3.53LSLQTTHH193 pKa = 6.35QFLDD197 pKa = 4.54AIARR201 pKa = 11.84MMEE204 pKa = 3.69NSRR207 pKa = 11.84WVVTNLPRR215 pKa = 11.84EE216 pKa = 4.3AYY218 pKa = 9.57SRR220 pKa = 11.84IYY222 pKa = 10.87GGLQNYY228 pKa = 7.98YY229 pKa = 11.09AEE231 pKa = 4.8LPGINPAQRR240 pKa = 11.84RR241 pKa = 11.84QIEE244 pKa = 4.03RR245 pKa = 11.84ALEE248 pKa = 3.9YY249 pKa = 10.87SNRR252 pKa = 11.84PSIEE256 pKa = 4.13DD257 pKa = 3.37ANSRR261 pKa = 11.84QVLEE265 pKa = 4.66AEE267 pKa = 4.54LGRR270 pKa = 11.84PDD272 pKa = 3.34VQRR275 pKa = 11.84RR276 pKa = 11.84SQQQEE281 pKa = 4.04DD282 pKa = 3.78SSSWFEE288 pKa = 3.84SGANIMRR295 pKa = 11.84YY296 pKa = 8.22FAPGGAHH303 pKa = 6.28QRR305 pKa = 11.84VTPDD309 pKa = 2.33WMLPLILGLYY319 pKa = 10.31GDD321 pKa = 5.41ISPTWQTYY329 pKa = 8.91IDD331 pKa = 3.8EE332 pKa = 4.49VEE334 pKa = 4.13YY335 pKa = 11.09GPKK338 pKa = 9.45KK339 pKa = 9.75KK340 pKa = 9.97KK341 pKa = 10.14RR342 pKa = 11.84RR343 pKa = 11.84FQQ345 pKa = 3.43
MM1 pKa = 7.51GSAISVIIEE10 pKa = 4.14MISYY14 pKa = 10.09LSEE17 pKa = 3.78ISSVTGISVEE27 pKa = 4.46AILSGEE33 pKa = 3.91AFAAIDD39 pKa = 3.7AQVTSLITMEE49 pKa = 4.51GFLGAEE55 pKa = 4.29TALSSIGLSEE65 pKa = 4.76DD66 pKa = 3.07MFIFMQAAPEE76 pKa = 4.07LTSTVMTEE84 pKa = 4.26FVRR87 pKa = 11.84EE88 pKa = 4.16SVQTAFIFQTVAGSAAFSLGSLHH111 pKa = 7.19GYY113 pKa = 8.7LAHH116 pKa = 6.8EE117 pKa = 4.35VPIVNRR123 pKa = 11.84NMALIPRR130 pKa = 11.84RR131 pKa = 11.84PADD134 pKa = 3.37YY135 pKa = 11.21YY136 pKa = 11.28DD137 pKa = 3.24ILFPGVQSFTHH148 pKa = 6.32ALDD151 pKa = 4.59VIHH154 pKa = 6.89GWGHH158 pKa = 6.21SLFQSVGEE166 pKa = 4.51YY167 pKa = 9.89IWDD170 pKa = 3.59TLRR173 pKa = 11.84RR174 pKa = 11.84EE175 pKa = 4.04TQGAVEE181 pKa = 4.18SAVRR185 pKa = 11.84DD186 pKa = 3.53LSLQTTHH193 pKa = 6.35QFLDD197 pKa = 4.54AIARR201 pKa = 11.84MMEE204 pKa = 3.69NSRR207 pKa = 11.84WVVTNLPRR215 pKa = 11.84EE216 pKa = 4.3AYY218 pKa = 9.57SRR220 pKa = 11.84IYY222 pKa = 10.87GGLQNYY228 pKa = 7.98YY229 pKa = 11.09AEE231 pKa = 4.8LPGINPAQRR240 pKa = 11.84RR241 pKa = 11.84QIEE244 pKa = 4.03RR245 pKa = 11.84ALEE248 pKa = 3.9YY249 pKa = 10.87SNRR252 pKa = 11.84PSIEE256 pKa = 4.13DD257 pKa = 3.37ANSRR261 pKa = 11.84QVLEE265 pKa = 4.66AEE267 pKa = 4.54LGRR270 pKa = 11.84PDD272 pKa = 3.34VQRR275 pKa = 11.84RR276 pKa = 11.84SQQQEE281 pKa = 4.04DD282 pKa = 3.78SSSWFEE288 pKa = 3.84SGANIMRR295 pKa = 11.84YY296 pKa = 8.22FAPGGAHH303 pKa = 6.28QRR305 pKa = 11.84VTPDD309 pKa = 2.33WMLPLILGLYY319 pKa = 10.31GDD321 pKa = 5.41ISPTWQTYY329 pKa = 8.91IDD331 pKa = 3.8EE332 pKa = 4.49VEE334 pKa = 4.13YY335 pKa = 11.09GPKK338 pKa = 9.45KK339 pKa = 9.75KK340 pKa = 9.97KK341 pKa = 10.14RR342 pKa = 11.84RR343 pKa = 11.84FQQ345 pKa = 3.43
Molecular weight: 38.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03080|ST_POVHA Small t antigen OS=Hamster polyomavirus OX=1891729 PE=3 SV=1
MM1 pKa = 7.89DD2 pKa = 6.22RR3 pKa = 11.84ILTKK7 pKa = 9.85EE8 pKa = 4.18EE9 pKa = 3.69KK10 pKa = 9.88QALISLLDD18 pKa = 4.39LEE20 pKa = 4.3PQYY23 pKa = 10.79WGDD26 pKa = 3.48YY27 pKa = 11.05GRR29 pKa = 11.84MQKK32 pKa = 10.3CYY34 pKa = 10.37KK35 pKa = 10.05KK36 pKa = 10.45KK37 pKa = 10.28CLQLHH42 pKa = 6.36PDD44 pKa = 3.03KK45 pKa = 11.37GGNEE49 pKa = 3.96EE50 pKa = 6.16LMQQLNTLWTKK61 pKa = 10.95LKK63 pKa = 10.75DD64 pKa = 3.18GLYY67 pKa = 9.77RR68 pKa = 11.84VRR70 pKa = 11.84LLLGPSQVRR79 pKa = 11.84RR80 pKa = 11.84LGKK83 pKa = 9.84DD84 pKa = 2.79QWNLSLQQTFSGTYY98 pKa = 8.77FRR100 pKa = 11.84RR101 pKa = 11.84LCRR104 pKa = 11.84LPITCLRR111 pKa = 11.84NKK113 pKa = 10.41GISTCNCILCLLRR126 pKa = 11.84KK127 pKa = 8.81QHH129 pKa = 6.69FLLKK133 pKa = 10.37KK134 pKa = 8.75SWRR137 pKa = 11.84VPCLVLGEE145 pKa = 5.08CYY147 pKa = 10.58CIDD150 pKa = 5.31CFALWFGLPVTNMLVPLYY168 pKa = 10.31AQFLAPIPVDD178 pKa = 3.1WLDD181 pKa = 4.15LNVHH185 pKa = 5.84EE186 pKa = 5.29VYY188 pKa = 10.96NPASGMTLMLPPPPADD204 pKa = 3.98PEE206 pKa = 4.34SSTILTQEE214 pKa = 4.03DD215 pKa = 3.87TGPTLMGQQDD225 pKa = 4.16TLTSRR230 pKa = 11.84RR231 pKa = 11.84NTGKK235 pKa = 10.31SFSLSGMLMRR245 pKa = 11.84TSPAKK250 pKa = 10.16KK251 pKa = 9.8SYY253 pKa = 9.77HH254 pKa = 4.81HH255 pKa = 6.48QKK257 pKa = 9.7MNSPPGIPIPPPPLFLFPVTAPVPPVTRR285 pKa = 11.84NTQEE289 pKa = 3.88TQAEE293 pKa = 4.29RR294 pKa = 11.84EE295 pKa = 4.04NEE297 pKa = 4.3YY298 pKa = 10.52MPMAPQIHH306 pKa = 7.42LYY308 pKa = 9.4SQIRR312 pKa = 11.84EE313 pKa = 4.13PTHH316 pKa = 6.39QEE318 pKa = 3.68EE319 pKa = 4.8EE320 pKa = 4.21EE321 pKa = 4.15PQYY324 pKa = 11.37EE325 pKa = 4.44EE326 pKa = 3.79IPIYY330 pKa = 10.95LEE332 pKa = 4.32LLPEE336 pKa = 4.49NPNQHH341 pKa = 6.8LALTSTARR349 pKa = 11.84RR350 pKa = 11.84SLRR353 pKa = 11.84RR354 pKa = 11.84KK355 pKa = 6.76YY356 pKa = 10.04HH357 pKa = 5.32KK358 pKa = 10.1HH359 pKa = 5.24NSHH362 pKa = 7.6IITQRR367 pKa = 11.84QRR369 pKa = 11.84NRR371 pKa = 11.84LRR373 pKa = 11.84RR374 pKa = 11.84LVLMIFLLSLGGFFLTLFFLIKK396 pKa = 10.35RR397 pKa = 11.84KK398 pKa = 8.21MHH400 pKa = 6.39LL401 pKa = 3.49
MM1 pKa = 7.89DD2 pKa = 6.22RR3 pKa = 11.84ILTKK7 pKa = 9.85EE8 pKa = 4.18EE9 pKa = 3.69KK10 pKa = 9.88QALISLLDD18 pKa = 4.39LEE20 pKa = 4.3PQYY23 pKa = 10.79WGDD26 pKa = 3.48YY27 pKa = 11.05GRR29 pKa = 11.84MQKK32 pKa = 10.3CYY34 pKa = 10.37KK35 pKa = 10.05KK36 pKa = 10.45KK37 pKa = 10.28CLQLHH42 pKa = 6.36PDD44 pKa = 3.03KK45 pKa = 11.37GGNEE49 pKa = 3.96EE50 pKa = 6.16LMQQLNTLWTKK61 pKa = 10.95LKK63 pKa = 10.75DD64 pKa = 3.18GLYY67 pKa = 9.77RR68 pKa = 11.84VRR70 pKa = 11.84LLLGPSQVRR79 pKa = 11.84RR80 pKa = 11.84LGKK83 pKa = 9.84DD84 pKa = 2.79QWNLSLQQTFSGTYY98 pKa = 8.77FRR100 pKa = 11.84RR101 pKa = 11.84LCRR104 pKa = 11.84LPITCLRR111 pKa = 11.84NKK113 pKa = 10.41GISTCNCILCLLRR126 pKa = 11.84KK127 pKa = 8.81QHH129 pKa = 6.69FLLKK133 pKa = 10.37KK134 pKa = 8.75SWRR137 pKa = 11.84VPCLVLGEE145 pKa = 5.08CYY147 pKa = 10.58CIDD150 pKa = 5.31CFALWFGLPVTNMLVPLYY168 pKa = 10.31AQFLAPIPVDD178 pKa = 3.1WLDD181 pKa = 4.15LNVHH185 pKa = 5.84EE186 pKa = 5.29VYY188 pKa = 10.96NPASGMTLMLPPPPADD204 pKa = 3.98PEE206 pKa = 4.34SSTILTQEE214 pKa = 4.03DD215 pKa = 3.87TGPTLMGQQDD225 pKa = 4.16TLTSRR230 pKa = 11.84RR231 pKa = 11.84NTGKK235 pKa = 10.31SFSLSGMLMRR245 pKa = 11.84TSPAKK250 pKa = 10.16KK251 pKa = 9.8SYY253 pKa = 9.77HH254 pKa = 4.81HH255 pKa = 6.48QKK257 pKa = 9.7MNSPPGIPIPPPPLFLFPVTAPVPPVTRR285 pKa = 11.84NTQEE289 pKa = 3.88TQAEE293 pKa = 4.29RR294 pKa = 11.84EE295 pKa = 4.04NEE297 pKa = 4.3YY298 pKa = 10.52MPMAPQIHH306 pKa = 7.42LYY308 pKa = 9.4SQIRR312 pKa = 11.84EE313 pKa = 4.13PTHH316 pKa = 6.39QEE318 pKa = 3.68EE319 pKa = 4.8EE320 pKa = 4.21EE321 pKa = 4.15PQYY324 pKa = 11.37EE325 pKa = 4.44EE326 pKa = 3.79IPIYY330 pKa = 10.95LEE332 pKa = 4.32LLPEE336 pKa = 4.49NPNQHH341 pKa = 6.8LALTSTARR349 pKa = 11.84RR350 pKa = 11.84SLRR353 pKa = 11.84RR354 pKa = 11.84KK355 pKa = 6.76YY356 pKa = 10.04HH357 pKa = 5.32KK358 pKa = 10.1HH359 pKa = 5.24NSHH362 pKa = 7.6IITQRR367 pKa = 11.84QRR369 pKa = 11.84NRR371 pKa = 11.84LRR373 pKa = 11.84RR374 pKa = 11.84LVLMIFLLSLGGFFLTLFFLIKK396 pKa = 10.35RR397 pKa = 11.84KK398 pKa = 8.21MHH400 pKa = 6.39LL401 pKa = 3.49
Molecular weight: 46.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2284 |
194 |
751 |
380.7 |
42.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.429 ± 0.821 | 2.145 ± 0.654 |
4.641 ± 0.448 | 6.305 ± 0.522 |
3.722 ± 0.589 | 6.13 ± 0.787 |
2.189 ± 0.282 | 4.51 ± 0.644 |
5.648 ± 1.197 | 10.814 ± 1.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.671 ± 0.263 | 4.466 ± 0.514 |
6.173 ± 0.586 | 5.385 ± 0.419 |
5.473 ± 0.834 | 6.524 ± 0.75 |
5.648 ± 0.48 | 5.385 ± 0.601 |
1.751 ± 0.23 | 4.072 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
