
Seal anellovirus 6
Taxonomy: Viruses; Anelloviridae; Lambdatorquevirus; Torque teno pinniped virus 3
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
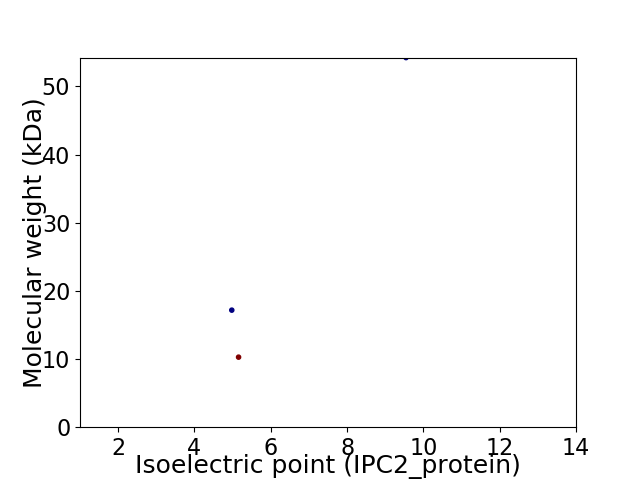
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7TWN7|A0A0A7TWN7_9VIRU Capsid protein OS=Seal anellovirus 6 OX=1566012 PE=3 SV=1
MM1 pKa = 7.22TKK3 pKa = 9.64SHH5 pKa = 6.56SGFAIVSNLRR15 pKa = 11.84SLEE18 pKa = 4.11TLSTEE23 pKa = 4.16DD24 pKa = 3.43YY25 pKa = 10.22RR26 pKa = 11.84RR27 pKa = 11.84SAIQTPSLKK36 pKa = 10.42HH37 pKa = 5.51QEE39 pKa = 4.2LKK41 pKa = 9.59TKK43 pKa = 10.43YY44 pKa = 9.95RR45 pKa = 11.84LAPYY49 pKa = 10.36SKK51 pKa = 10.89LDD53 pKa = 3.57LSTLGTYY60 pKa = 10.81LKK62 pKa = 9.86GTSRR66 pKa = 11.84TAASRR71 pKa = 11.84KK72 pKa = 9.14RR73 pKa = 11.84VLEE76 pKa = 3.87EE77 pKa = 3.77LLDD80 pKa = 4.76LIEE83 pKa = 5.76EE84 pKa = 4.47DD85 pKa = 5.1SQPNWQTQAQDD96 pKa = 3.19VSDD99 pKa = 4.88FSTTPGSEE107 pKa = 4.12SEE109 pKa = 4.3EE110 pKa = 4.34SDD112 pKa = 3.85VSYY115 pKa = 11.0LGYY118 pKa = 10.14WSDD121 pKa = 3.69EE122 pKa = 4.11EE123 pKa = 4.57NPGGEE128 pKa = 4.57GPPLVPPTPGPKK140 pKa = 9.67APEE143 pKa = 4.12YY144 pKa = 9.67PPHH147 pKa = 6.74FLFLTLQQ154 pKa = 3.44
MM1 pKa = 7.22TKK3 pKa = 9.64SHH5 pKa = 6.56SGFAIVSNLRR15 pKa = 11.84SLEE18 pKa = 4.11TLSTEE23 pKa = 4.16DD24 pKa = 3.43YY25 pKa = 10.22RR26 pKa = 11.84RR27 pKa = 11.84SAIQTPSLKK36 pKa = 10.42HH37 pKa = 5.51QEE39 pKa = 4.2LKK41 pKa = 9.59TKK43 pKa = 10.43YY44 pKa = 9.95RR45 pKa = 11.84LAPYY49 pKa = 10.36SKK51 pKa = 10.89LDD53 pKa = 3.57LSTLGTYY60 pKa = 10.81LKK62 pKa = 9.86GTSRR66 pKa = 11.84TAASRR71 pKa = 11.84KK72 pKa = 9.14RR73 pKa = 11.84VLEE76 pKa = 3.87EE77 pKa = 3.77LLDD80 pKa = 4.76LIEE83 pKa = 5.76EE84 pKa = 4.47DD85 pKa = 5.1SQPNWQTQAQDD96 pKa = 3.19VSDD99 pKa = 4.88FSTTPGSEE107 pKa = 4.12SEE109 pKa = 4.3EE110 pKa = 4.34SDD112 pKa = 3.85VSYY115 pKa = 11.0LGYY118 pKa = 10.14WSDD121 pKa = 3.69EE122 pKa = 4.11EE123 pKa = 4.57NPGGEE128 pKa = 4.57GPPLVPPTPGPKK140 pKa = 9.67APEE143 pKa = 4.12YY144 pKa = 9.67PPHH147 pKa = 6.74FLFLTLQQ154 pKa = 3.44
Molecular weight: 17.18 kDa
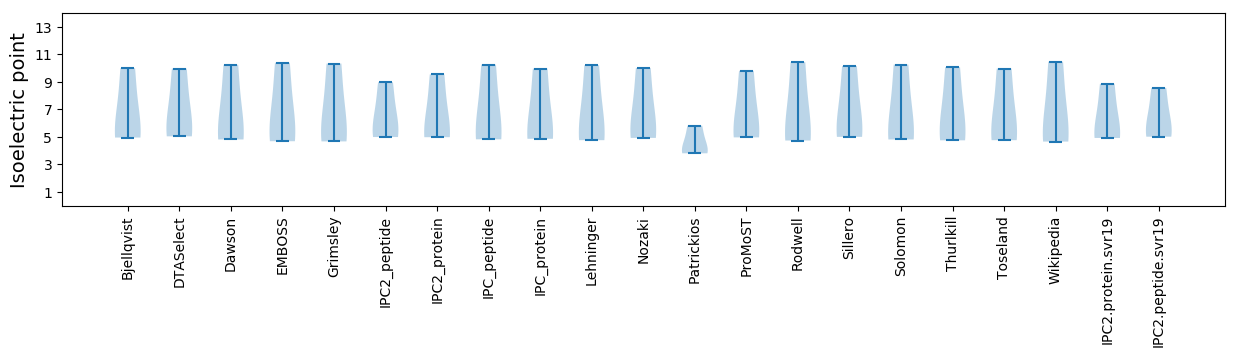
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7U2J0|A0A0A7U2J0_9VIRU ORF2 OS=Seal anellovirus 6 OX=1566012 PE=4 SV=1
MM1 pKa = 8.06AYY3 pKa = 9.86RR4 pKa = 11.84HH5 pKa = 5.89RR6 pKa = 11.84RR7 pKa = 11.84PFRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 8.17RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.05PRR18 pKa = 11.84WKK20 pKa = 9.02GRR22 pKa = 11.84RR23 pKa = 11.84VHH25 pKa = 6.89RR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 4.57TWRR31 pKa = 11.84PFHH34 pKa = 6.37HH35 pKa = 5.93WRR37 pKa = 11.84RR38 pKa = 11.84WRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PYY44 pKa = 10.28RR45 pKa = 11.84RR46 pKa = 11.84GTVAVRR52 pKa = 11.84YY53 pKa = 6.78TNPRR57 pKa = 11.84RR58 pKa = 11.84HH59 pKa = 6.27KK60 pKa = 9.57YY61 pKa = 9.98ISVTGWEE68 pKa = 4.29PLGNCCIQDD77 pKa = 3.64KK78 pKa = 10.97ASDD81 pKa = 3.62EE82 pKa = 4.1ATPYY86 pKa = 10.97EE87 pKa = 4.74DD88 pKa = 5.44LDD90 pKa = 4.5SDD92 pKa = 4.05VQTWANRR99 pKa = 11.84TRR101 pKa = 11.84TAGIWHH107 pKa = 6.55GTYY110 pKa = 9.59GHH112 pKa = 7.25HH113 pKa = 6.74YY114 pKa = 8.98PWQLLCQRR122 pKa = 11.84AQFYY126 pKa = 10.62FCTFSSGWKK135 pKa = 10.23GYY137 pKa = 10.82DD138 pKa = 3.13YY139 pKa = 11.25LQFYY143 pKa = 10.05GGNIWLPKK151 pKa = 9.62LAAPYY156 pKa = 8.73FFWVDD161 pKa = 3.42PTVQDD166 pKa = 4.03PKK168 pKa = 10.34KK169 pKa = 10.4SSYY172 pKa = 10.87EE173 pKa = 4.11SLCEE177 pKa = 4.19GQDD180 pKa = 3.51LVPPRR185 pKa = 11.84GIHH188 pKa = 4.62EE189 pKa = 4.39QKK191 pKa = 9.93GAHH194 pKa = 6.18YY195 pKa = 10.1IPANNEE201 pKa = 3.53SHH203 pKa = 5.66TLKK206 pKa = 10.42GYY208 pKa = 10.45KK209 pKa = 9.49RR210 pKa = 11.84IHH212 pKa = 6.62IKK214 pKa = 10.5RR215 pKa = 11.84LSTWEE220 pKa = 3.77GVYY223 pKa = 10.56AISTAADD230 pKa = 3.82YY231 pKa = 11.22ILVHH235 pKa = 5.98WVWSTVTKK243 pKa = 10.65NWAFFDD249 pKa = 4.43GACYY253 pKa = 9.7QAGQASSDD261 pKa = 3.96TCNSTPWFMSAQTAEE276 pKa = 4.23SGVKK280 pKa = 10.24KK281 pKa = 10.57EE282 pKa = 3.48VDD284 pKa = 3.49EE285 pKa = 4.28YY286 pKa = 11.8KK287 pKa = 9.66KK288 pKa = 9.53TNDD291 pKa = 3.44FKK293 pKa = 11.82NNLYY297 pKa = 10.4CYY299 pKa = 8.62PDD301 pKa = 3.97PRR303 pKa = 11.84ASWVNRR309 pKa = 11.84QKK311 pKa = 11.16YY312 pKa = 9.91NKK314 pKa = 8.6TIQPLTVTEE323 pKa = 4.22NPGGPFLPSQGAGLGLSRR341 pKa = 11.84QYY343 pKa = 11.25DD344 pKa = 3.34QVSFWFRR351 pKa = 11.84YY352 pKa = 8.16RR353 pKa = 11.84FKK355 pKa = 10.94FKK357 pKa = 10.99VSGDD361 pKa = 3.35SVYY364 pKa = 10.76RR365 pKa = 11.84RR366 pKa = 11.84LPAVSNTDD374 pKa = 4.09TIPEE378 pKa = 4.04APGAQNQVSPRR389 pKa = 11.84SILKK393 pKa = 10.04ARR395 pKa = 11.84PLDD398 pKa = 3.59TGDD401 pKa = 3.54ILEE404 pKa = 5.3GDD406 pKa = 4.12LQDD409 pKa = 3.62GCLTEE414 pKa = 3.98EE415 pKa = 4.06SLRR418 pKa = 11.84RR419 pKa = 11.84ITGSHH424 pKa = 6.24RR425 pKa = 11.84RR426 pKa = 11.84RR427 pKa = 11.84LPTKK431 pKa = 10.27LADD434 pKa = 3.41SSSRR438 pKa = 11.84RR439 pKa = 11.84VRR441 pKa = 11.84LFHH444 pKa = 7.55DD445 pKa = 3.58PGVRR449 pKa = 11.84KK450 pKa = 9.73RR451 pKa = 11.84RR452 pKa = 11.84IRR454 pKa = 11.84RR455 pKa = 11.84LLSGLLEE462 pKa = 4.23
MM1 pKa = 8.06AYY3 pKa = 9.86RR4 pKa = 11.84HH5 pKa = 5.89RR6 pKa = 11.84RR7 pKa = 11.84PFRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 8.17RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.05PRR18 pKa = 11.84WKK20 pKa = 9.02GRR22 pKa = 11.84RR23 pKa = 11.84VHH25 pKa = 6.89RR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 4.57TWRR31 pKa = 11.84PFHH34 pKa = 6.37HH35 pKa = 5.93WRR37 pKa = 11.84RR38 pKa = 11.84WRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PYY44 pKa = 10.28RR45 pKa = 11.84RR46 pKa = 11.84GTVAVRR52 pKa = 11.84YY53 pKa = 6.78TNPRR57 pKa = 11.84RR58 pKa = 11.84HH59 pKa = 6.27KK60 pKa = 9.57YY61 pKa = 9.98ISVTGWEE68 pKa = 4.29PLGNCCIQDD77 pKa = 3.64KK78 pKa = 10.97ASDD81 pKa = 3.62EE82 pKa = 4.1ATPYY86 pKa = 10.97EE87 pKa = 4.74DD88 pKa = 5.44LDD90 pKa = 4.5SDD92 pKa = 4.05VQTWANRR99 pKa = 11.84TRR101 pKa = 11.84TAGIWHH107 pKa = 6.55GTYY110 pKa = 9.59GHH112 pKa = 7.25HH113 pKa = 6.74YY114 pKa = 8.98PWQLLCQRR122 pKa = 11.84AQFYY126 pKa = 10.62FCTFSSGWKK135 pKa = 10.23GYY137 pKa = 10.82DD138 pKa = 3.13YY139 pKa = 11.25LQFYY143 pKa = 10.05GGNIWLPKK151 pKa = 9.62LAAPYY156 pKa = 8.73FFWVDD161 pKa = 3.42PTVQDD166 pKa = 4.03PKK168 pKa = 10.34KK169 pKa = 10.4SSYY172 pKa = 10.87EE173 pKa = 4.11SLCEE177 pKa = 4.19GQDD180 pKa = 3.51LVPPRR185 pKa = 11.84GIHH188 pKa = 4.62EE189 pKa = 4.39QKK191 pKa = 9.93GAHH194 pKa = 6.18YY195 pKa = 10.1IPANNEE201 pKa = 3.53SHH203 pKa = 5.66TLKK206 pKa = 10.42GYY208 pKa = 10.45KK209 pKa = 9.49RR210 pKa = 11.84IHH212 pKa = 6.62IKK214 pKa = 10.5RR215 pKa = 11.84LSTWEE220 pKa = 3.77GVYY223 pKa = 10.56AISTAADD230 pKa = 3.82YY231 pKa = 11.22ILVHH235 pKa = 5.98WVWSTVTKK243 pKa = 10.65NWAFFDD249 pKa = 4.43GACYY253 pKa = 9.7QAGQASSDD261 pKa = 3.96TCNSTPWFMSAQTAEE276 pKa = 4.23SGVKK280 pKa = 10.24KK281 pKa = 10.57EE282 pKa = 3.48VDD284 pKa = 3.49EE285 pKa = 4.28YY286 pKa = 11.8KK287 pKa = 9.66KK288 pKa = 9.53TNDD291 pKa = 3.44FKK293 pKa = 11.82NNLYY297 pKa = 10.4CYY299 pKa = 8.62PDD301 pKa = 3.97PRR303 pKa = 11.84ASWVNRR309 pKa = 11.84QKK311 pKa = 11.16YY312 pKa = 9.91NKK314 pKa = 8.6TIQPLTVTEE323 pKa = 4.22NPGGPFLPSQGAGLGLSRR341 pKa = 11.84QYY343 pKa = 11.25DD344 pKa = 3.34QVSFWFRR351 pKa = 11.84YY352 pKa = 8.16RR353 pKa = 11.84FKK355 pKa = 10.94FKK357 pKa = 10.99VSGDD361 pKa = 3.35SVYY364 pKa = 10.76RR365 pKa = 11.84RR366 pKa = 11.84LPAVSNTDD374 pKa = 4.09TIPEE378 pKa = 4.04APGAQNQVSPRR389 pKa = 11.84SILKK393 pKa = 10.04ARR395 pKa = 11.84PLDD398 pKa = 3.59TGDD401 pKa = 3.54ILEE404 pKa = 5.3GDD406 pKa = 4.12LQDD409 pKa = 3.62GCLTEE414 pKa = 3.98EE415 pKa = 4.06SLRR418 pKa = 11.84RR419 pKa = 11.84ITGSHH424 pKa = 6.24RR425 pKa = 11.84RR426 pKa = 11.84RR427 pKa = 11.84LPTKK431 pKa = 10.27LADD434 pKa = 3.41SSSRR438 pKa = 11.84RR439 pKa = 11.84VRR441 pKa = 11.84LFHH444 pKa = 7.55DD445 pKa = 3.58PGVRR449 pKa = 11.84KK450 pKa = 9.73RR451 pKa = 11.84RR452 pKa = 11.84IRR454 pKa = 11.84RR455 pKa = 11.84LLSGLLEE462 pKa = 4.23
Molecular weight: 54.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
714 |
98 |
462 |
238.0 |
27.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.742 ± 0.516 | 1.681 ± 0.672 |
5.602 ± 0.652 | 5.042 ± 1.666 |
3.501 ± 0.423 | 7.983 ± 2.126 |
3.501 ± 0.785 | 2.941 ± 0.534 |
5.182 ± 0.02 | 8.123 ± 1.732 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.56 ± 0.139 | 3.081 ± 0.466 |
6.723 ± 0.931 | 4.342 ± 0.09 |
8.543 ± 2.739 | 8.123 ± 1.665 |
7.283 ± 1.022 | 4.062 ± 0.971 |
3.081 ± 0.86 | 4.902 ± 1.093 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
