
Klebsiella phage K64-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Alcyoneusvirus; Klebsiella virus K64-1
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
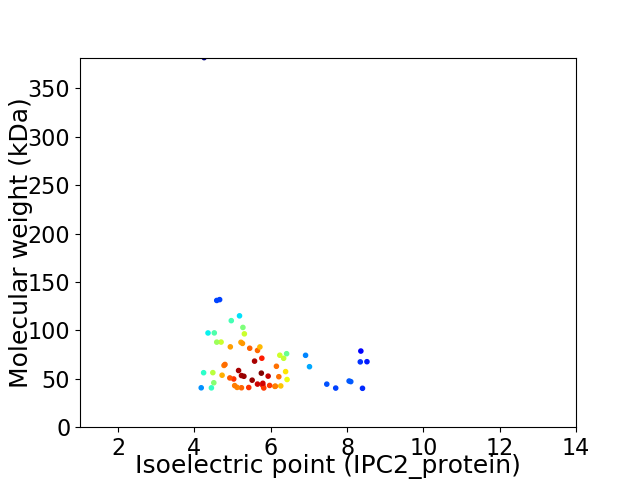
Virtual 2D-PAGE plot for 64 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A8J9R3|A0A0A8J9R3_9CAUD Uncharacterized protein OS=Klebsiella phage K64-1 OX=1439894 PE=4 SV=1
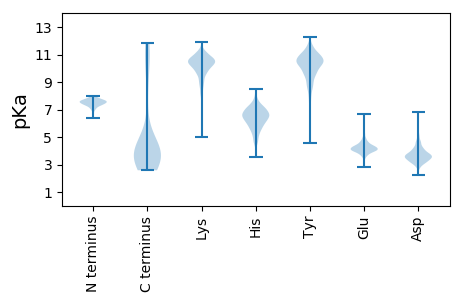
MM1 pKa = 7.59TISFNNGSNCSCNNSGIKK19 pKa = 9.92SMKK22 pKa = 9.81ISTDD26 pKa = 2.29GHH28 pKa = 6.85LIVTYY33 pKa = 10.87ADD35 pKa = 3.38NTIEE39 pKa = 4.68DD40 pKa = 3.74LGLVVGHH47 pKa = 7.25DD48 pKa = 4.03GSDD51 pKa = 3.22GSNFYY56 pKa = 10.49PNEE59 pKa = 3.93IGFEE63 pKa = 4.06IPDD66 pKa = 3.55SDD68 pKa = 4.22FNTDD72 pKa = 3.26KK73 pKa = 11.11TIGWSYY79 pKa = 11.41LSLANSTSVLYY90 pKa = 10.26FKK92 pKa = 10.26TSNPGEE98 pKa = 4.14SSTWNSVEE106 pKa = 4.11FGKK109 pKa = 11.04GEE111 pKa = 3.81QGEE114 pKa = 4.35QGKK117 pKa = 8.62PFHH120 pKa = 7.12IDD122 pKa = 2.48SSGTVFPTSGLFDD135 pKa = 4.66GYY137 pKa = 9.47TFYY140 pKa = 10.67NTNDD144 pKa = 3.27GKK146 pKa = 11.35LYY148 pKa = 10.32FYY150 pKa = 10.89DD151 pKa = 4.74SSTQTWSSGISFRR164 pKa = 11.84GEE166 pKa = 3.43VGPRR170 pKa = 11.84GRR172 pKa = 11.84FHH174 pKa = 7.4IDD176 pKa = 2.86STGSTFPDD184 pKa = 3.07ITNLPEE190 pKa = 5.22DD191 pKa = 3.73YY192 pKa = 10.73CFYY195 pKa = 10.09NTSDD199 pKa = 3.01GKK201 pKa = 10.89LYY203 pKa = 10.98YY204 pKa = 10.4VDD206 pKa = 3.75IVEE209 pKa = 4.51NTDD212 pKa = 3.47PVQKK216 pKa = 9.97EE217 pKa = 3.78WSTGIEE223 pKa = 3.97FRR225 pKa = 11.84GKK227 pKa = 10.45EE228 pKa = 3.97GLQGEE233 pKa = 4.62KK234 pKa = 10.38GDD236 pKa = 4.66KK237 pKa = 10.77GDD239 pKa = 3.64AGKK242 pKa = 10.51DD243 pKa = 2.75IVVIVNTIDD252 pKa = 3.09HH253 pKa = 6.87SYY255 pKa = 9.64TNTVLVIGTCPAGYY269 pKa = 9.92LVTNIEE275 pKa = 4.34VNVTQGYY282 pKa = 7.33EE283 pKa = 3.98PPVTDD288 pKa = 3.11MKK290 pKa = 10.91IRR292 pKa = 11.84FSGEE296 pKa = 3.54ANEE299 pKa = 5.45DD300 pKa = 3.03IGGTVIAGYY309 pKa = 10.55DD310 pKa = 3.48YY311 pKa = 11.4FDD313 pKa = 4.41ISMQSRR319 pKa = 11.84YY320 pKa = 10.25VINEE324 pKa = 3.88TNHH327 pKa = 5.52IVSDD331 pKa = 3.42KK332 pKa = 11.52DD333 pKa = 3.84EE334 pKa = 4.99IISCIFNEE342 pKa = 4.26SVNLSSYY349 pKa = 11.71GNMTIICTLSWQAPTLPISDD369 pKa = 4.65NII371 pKa = 3.84
MM1 pKa = 7.59TISFNNGSNCSCNNSGIKK19 pKa = 9.92SMKK22 pKa = 9.81ISTDD26 pKa = 2.29GHH28 pKa = 6.85LIVTYY33 pKa = 10.87ADD35 pKa = 3.38NTIEE39 pKa = 4.68DD40 pKa = 3.74LGLVVGHH47 pKa = 7.25DD48 pKa = 4.03GSDD51 pKa = 3.22GSNFYY56 pKa = 10.49PNEE59 pKa = 3.93IGFEE63 pKa = 4.06IPDD66 pKa = 3.55SDD68 pKa = 4.22FNTDD72 pKa = 3.26KK73 pKa = 11.11TIGWSYY79 pKa = 11.41LSLANSTSVLYY90 pKa = 10.26FKK92 pKa = 10.26TSNPGEE98 pKa = 4.14SSTWNSVEE106 pKa = 4.11FGKK109 pKa = 11.04GEE111 pKa = 3.81QGEE114 pKa = 4.35QGKK117 pKa = 8.62PFHH120 pKa = 7.12IDD122 pKa = 2.48SSGTVFPTSGLFDD135 pKa = 4.66GYY137 pKa = 9.47TFYY140 pKa = 10.67NTNDD144 pKa = 3.27GKK146 pKa = 11.35LYY148 pKa = 10.32FYY150 pKa = 10.89DD151 pKa = 4.74SSTQTWSSGISFRR164 pKa = 11.84GEE166 pKa = 3.43VGPRR170 pKa = 11.84GRR172 pKa = 11.84FHH174 pKa = 7.4IDD176 pKa = 2.86STGSTFPDD184 pKa = 3.07ITNLPEE190 pKa = 5.22DD191 pKa = 3.73YY192 pKa = 10.73CFYY195 pKa = 10.09NTSDD199 pKa = 3.01GKK201 pKa = 10.89LYY203 pKa = 10.98YY204 pKa = 10.4VDD206 pKa = 3.75IVEE209 pKa = 4.51NTDD212 pKa = 3.47PVQKK216 pKa = 9.97EE217 pKa = 3.78WSTGIEE223 pKa = 3.97FRR225 pKa = 11.84GKK227 pKa = 10.45EE228 pKa = 3.97GLQGEE233 pKa = 4.62KK234 pKa = 10.38GDD236 pKa = 4.66KK237 pKa = 10.77GDD239 pKa = 3.64AGKK242 pKa = 10.51DD243 pKa = 2.75IVVIVNTIDD252 pKa = 3.09HH253 pKa = 6.87SYY255 pKa = 9.64TNTVLVIGTCPAGYY269 pKa = 9.92LVTNIEE275 pKa = 4.34VNVTQGYY282 pKa = 7.33EE283 pKa = 3.98PPVTDD288 pKa = 3.11MKK290 pKa = 10.91IRR292 pKa = 11.84FSGEE296 pKa = 3.54ANEE299 pKa = 5.45DD300 pKa = 3.03IGGTVIAGYY309 pKa = 10.55DD310 pKa = 3.48YY311 pKa = 11.4FDD313 pKa = 4.41ISMQSRR319 pKa = 11.84YY320 pKa = 10.25VINEE324 pKa = 3.88TNHH327 pKa = 5.52IVSDD331 pKa = 3.42KK332 pKa = 11.52DD333 pKa = 3.84EE334 pKa = 4.99IISCIFNEE342 pKa = 4.26SVNLSSYY349 pKa = 11.71GNMTIICTLSWQAPTLPISDD369 pKa = 4.65NII371 pKa = 3.84
Molecular weight: 40.69 kDa
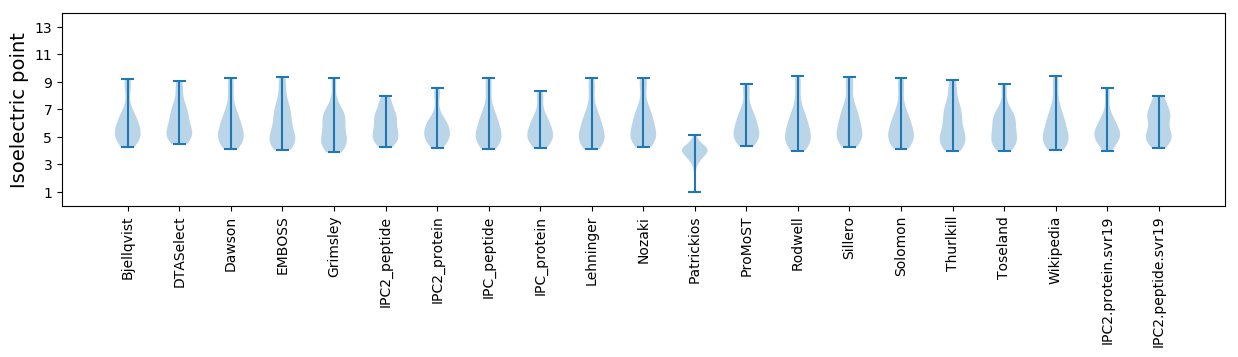
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A8J9Y6|A0A0A8J9Y6_9CAUD Putative Hef-like homing endonuclease OS=Klebsiella phage K64-1 OX=1439894 PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 10.25KK3 pKa = 10.15YY4 pKa = 10.74LKK6 pKa = 10.86NLDD9 pKa = 3.98TEE11 pKa = 4.59TEE13 pKa = 3.95NLFNEE18 pKa = 3.99IRR20 pKa = 11.84SLLSNKK26 pKa = 8.93NSKK29 pKa = 8.95SWHH32 pKa = 5.03TSINASGRR40 pKa = 11.84EE41 pKa = 3.94HH42 pKa = 7.07LKK44 pKa = 10.36NAVLRR49 pKa = 11.84VYY51 pKa = 10.33PSSDD55 pKa = 3.23FLLSEE60 pKa = 4.04ALKK63 pKa = 10.9LIINQEE69 pKa = 3.99EE70 pKa = 4.19PPYY73 pKa = 10.2AVCGRR78 pKa = 11.84KK79 pKa = 9.11CKK81 pKa = 10.48YY82 pKa = 10.66FNGQYY87 pKa = 10.87DD88 pKa = 4.5RR89 pKa = 11.84YY90 pKa = 9.78CSLGVKK96 pKa = 9.29PNKK99 pKa = 8.78NTYY102 pKa = 9.35CEE104 pKa = 4.14KK105 pKa = 10.73CNKK108 pKa = 8.89HH109 pKa = 4.65WRR111 pKa = 11.84EE112 pKa = 4.04KK113 pKa = 10.46KK114 pKa = 10.36NKK116 pKa = 7.45STKK119 pKa = 9.69EE120 pKa = 3.77NCLNQYY126 pKa = 9.48NVEE129 pKa = 4.47HH130 pKa = 7.0PFQRR134 pKa = 11.84NDD136 pKa = 2.87VKK138 pKa = 10.57TKK140 pKa = 8.68VHH142 pKa = 4.8EE143 pKa = 4.32TKK145 pKa = 9.83IRR147 pKa = 11.84KK148 pKa = 9.39YY149 pKa = 7.98GTNYY153 pKa = 10.67NKK155 pKa = 10.22IISNKK160 pKa = 9.97SFSTYY165 pKa = 9.74YY166 pKa = 10.86NKK168 pKa = 10.02TGYY171 pKa = 9.91SCPLSNPDD179 pKa = 2.88IQQKK183 pKa = 10.04IKK185 pKa = 8.99EE186 pKa = 4.59TNIKK190 pKa = 10.35NYY192 pKa = 10.27GYY194 pKa = 9.08EE195 pKa = 4.26NPGSSPLIVQKK206 pKa = 10.79RR207 pKa = 11.84KK208 pKa = 8.13NTQKK212 pKa = 10.83KK213 pKa = 10.15LYY215 pKa = 8.75GHH217 pKa = 6.02EE218 pKa = 4.51HH219 pKa = 6.04SMQNGDD225 pKa = 3.27IKK227 pKa = 11.18LKK229 pKa = 10.93VLIKK233 pKa = 10.63SATQRR238 pKa = 11.84MDD240 pKa = 3.9SINSKK245 pKa = 11.21LEE247 pKa = 3.69FTNFIPLFDD256 pKa = 4.01ISDD259 pKa = 4.89LIGLDD264 pKa = 3.84TYY266 pKa = 11.99NKK268 pKa = 8.92MPYY271 pKa = 8.25WCNTHH276 pKa = 6.27GLFYY280 pKa = 10.85SCSNYY285 pKa = 10.42KK286 pKa = 10.06IICNEE291 pKa = 3.98CTPVHH296 pKa = 6.97ISTQHH301 pKa = 6.46KK302 pKa = 9.81EE303 pKa = 3.54ILNFIEE309 pKa = 4.53NLGIEE314 pKa = 4.71CVTNTRR320 pKa = 11.84SVLKK324 pKa = 9.85STMEE328 pKa = 4.0LDD330 pKa = 4.09IFIPDD335 pKa = 3.7LNVAIEE341 pKa = 4.0INGMYY346 pKa = 8.8WHH348 pKa = 5.75STEE351 pKa = 4.03YY352 pKa = 10.67KK353 pKa = 9.84EE354 pKa = 5.69RR355 pKa = 11.84LYY357 pKa = 10.72HH358 pKa = 4.69QNKK361 pKa = 8.19MLEE364 pKa = 4.37CKK366 pKa = 9.68EE367 pKa = 4.17HH368 pKa = 6.84GIHH371 pKa = 7.97LIQIWEE377 pKa = 4.45DD378 pKa = 3.04EE379 pKa = 4.5WNTKK383 pKa = 8.96QDD385 pKa = 3.58LVKK388 pKa = 10.4FRR390 pKa = 11.84LMNVLKK396 pKa = 10.5CSSTIYY402 pKa = 10.56ARR404 pKa = 11.84KK405 pKa = 9.92CKK407 pKa = 10.08IKK409 pKa = 10.33EE410 pKa = 4.03INKK413 pKa = 9.63KK414 pKa = 10.06EE415 pKa = 3.85CDD417 pKa = 3.44NFINTYY423 pKa = 9.61HH424 pKa = 6.38IQGSSSSSIRR434 pKa = 11.84IGLYY438 pKa = 10.01YY439 pKa = 10.46EE440 pKa = 4.22NEE442 pKa = 4.29LYY444 pKa = 11.04AVMTFGKK451 pKa = 10.32SRR453 pKa = 11.84FDD455 pKa = 3.1KK456 pKa = 9.88TVQYY460 pKa = 11.04EE461 pKa = 4.25LIRR464 pKa = 11.84YY465 pKa = 7.4CSKK468 pKa = 9.64STVVGGASKK477 pKa = 10.58LLKK480 pKa = 10.65YY481 pKa = 10.24FEE483 pKa = 4.73KK484 pKa = 10.39KK485 pKa = 10.02YY486 pKa = 10.88KK487 pKa = 9.42PVSLISYY494 pKa = 10.9ADD496 pKa = 3.8ACWSTGDD503 pKa = 4.66LYY505 pKa = 10.78IKK507 pKa = 10.77LGFTYY512 pKa = 10.7NGLTEE517 pKa = 4.5PGYY520 pKa = 10.44FYY522 pKa = 11.19YY523 pKa = 10.56KK524 pKa = 10.41SSTGRR529 pKa = 11.84ISRR532 pKa = 11.84HH533 pKa = 3.54QCQKK537 pKa = 11.01KK538 pKa = 9.71KK539 pKa = 10.64LVNQGYY545 pKa = 10.15DD546 pKa = 3.43EE547 pKa = 4.64NLSEE551 pKa = 4.16YY552 pKa = 10.25QICTTVLKK560 pKa = 10.45YY561 pKa = 11.18NKK563 pKa = 9.77IFDD566 pKa = 4.04CGNFKK571 pKa = 9.11FTKK574 pKa = 10.39YY575 pKa = 10.77YY576 pKa = 9.77II577 pKa = 3.9
MM1 pKa = 7.71KK2 pKa = 10.25KK3 pKa = 10.15YY4 pKa = 10.74LKK6 pKa = 10.86NLDD9 pKa = 3.98TEE11 pKa = 4.59TEE13 pKa = 3.95NLFNEE18 pKa = 3.99IRR20 pKa = 11.84SLLSNKK26 pKa = 8.93NSKK29 pKa = 8.95SWHH32 pKa = 5.03TSINASGRR40 pKa = 11.84EE41 pKa = 3.94HH42 pKa = 7.07LKK44 pKa = 10.36NAVLRR49 pKa = 11.84VYY51 pKa = 10.33PSSDD55 pKa = 3.23FLLSEE60 pKa = 4.04ALKK63 pKa = 10.9LIINQEE69 pKa = 3.99EE70 pKa = 4.19PPYY73 pKa = 10.2AVCGRR78 pKa = 11.84KK79 pKa = 9.11CKK81 pKa = 10.48YY82 pKa = 10.66FNGQYY87 pKa = 10.87DD88 pKa = 4.5RR89 pKa = 11.84YY90 pKa = 9.78CSLGVKK96 pKa = 9.29PNKK99 pKa = 8.78NTYY102 pKa = 9.35CEE104 pKa = 4.14KK105 pKa = 10.73CNKK108 pKa = 8.89HH109 pKa = 4.65WRR111 pKa = 11.84EE112 pKa = 4.04KK113 pKa = 10.46KK114 pKa = 10.36NKK116 pKa = 7.45STKK119 pKa = 9.69EE120 pKa = 3.77NCLNQYY126 pKa = 9.48NVEE129 pKa = 4.47HH130 pKa = 7.0PFQRR134 pKa = 11.84NDD136 pKa = 2.87VKK138 pKa = 10.57TKK140 pKa = 8.68VHH142 pKa = 4.8EE143 pKa = 4.32TKK145 pKa = 9.83IRR147 pKa = 11.84KK148 pKa = 9.39YY149 pKa = 7.98GTNYY153 pKa = 10.67NKK155 pKa = 10.22IISNKK160 pKa = 9.97SFSTYY165 pKa = 9.74YY166 pKa = 10.86NKK168 pKa = 10.02TGYY171 pKa = 9.91SCPLSNPDD179 pKa = 2.88IQQKK183 pKa = 10.04IKK185 pKa = 8.99EE186 pKa = 4.59TNIKK190 pKa = 10.35NYY192 pKa = 10.27GYY194 pKa = 9.08EE195 pKa = 4.26NPGSSPLIVQKK206 pKa = 10.79RR207 pKa = 11.84KK208 pKa = 8.13NTQKK212 pKa = 10.83KK213 pKa = 10.15LYY215 pKa = 8.75GHH217 pKa = 6.02EE218 pKa = 4.51HH219 pKa = 6.04SMQNGDD225 pKa = 3.27IKK227 pKa = 11.18LKK229 pKa = 10.93VLIKK233 pKa = 10.63SATQRR238 pKa = 11.84MDD240 pKa = 3.9SINSKK245 pKa = 11.21LEE247 pKa = 3.69FTNFIPLFDD256 pKa = 4.01ISDD259 pKa = 4.89LIGLDD264 pKa = 3.84TYY266 pKa = 11.99NKK268 pKa = 8.92MPYY271 pKa = 8.25WCNTHH276 pKa = 6.27GLFYY280 pKa = 10.85SCSNYY285 pKa = 10.42KK286 pKa = 10.06IICNEE291 pKa = 3.98CTPVHH296 pKa = 6.97ISTQHH301 pKa = 6.46KK302 pKa = 9.81EE303 pKa = 3.54ILNFIEE309 pKa = 4.53NLGIEE314 pKa = 4.71CVTNTRR320 pKa = 11.84SVLKK324 pKa = 9.85STMEE328 pKa = 4.0LDD330 pKa = 4.09IFIPDD335 pKa = 3.7LNVAIEE341 pKa = 4.0INGMYY346 pKa = 8.8WHH348 pKa = 5.75STEE351 pKa = 4.03YY352 pKa = 10.67KK353 pKa = 9.84EE354 pKa = 5.69RR355 pKa = 11.84LYY357 pKa = 10.72HH358 pKa = 4.69QNKK361 pKa = 8.19MLEE364 pKa = 4.37CKK366 pKa = 9.68EE367 pKa = 4.17HH368 pKa = 6.84GIHH371 pKa = 7.97LIQIWEE377 pKa = 4.45DD378 pKa = 3.04EE379 pKa = 4.5WNTKK383 pKa = 8.96QDD385 pKa = 3.58LVKK388 pKa = 10.4FRR390 pKa = 11.84LMNVLKK396 pKa = 10.5CSSTIYY402 pKa = 10.56ARR404 pKa = 11.84KK405 pKa = 9.92CKK407 pKa = 10.08IKK409 pKa = 10.33EE410 pKa = 4.03INKK413 pKa = 9.63KK414 pKa = 10.06EE415 pKa = 3.85CDD417 pKa = 3.44NFINTYY423 pKa = 9.61HH424 pKa = 6.38IQGSSSSSIRR434 pKa = 11.84IGLYY438 pKa = 10.01YY439 pKa = 10.46EE440 pKa = 4.22NEE442 pKa = 4.29LYY444 pKa = 11.04AVMTFGKK451 pKa = 10.32SRR453 pKa = 11.84FDD455 pKa = 3.1KK456 pKa = 9.88TVQYY460 pKa = 11.04EE461 pKa = 4.25LIRR464 pKa = 11.84YY465 pKa = 7.4CSKK468 pKa = 9.64STVVGGASKK477 pKa = 10.58LLKK480 pKa = 10.65YY481 pKa = 10.24FEE483 pKa = 4.73KK484 pKa = 10.39KK485 pKa = 10.02YY486 pKa = 10.88KK487 pKa = 9.42PVSLISYY494 pKa = 10.9ADD496 pKa = 3.8ACWSTGDD503 pKa = 4.66LYY505 pKa = 10.78IKK507 pKa = 10.77LGFTYY512 pKa = 10.7NGLTEE517 pKa = 4.5PGYY520 pKa = 10.44FYY522 pKa = 11.19YY523 pKa = 10.56KK524 pKa = 10.41SSTGRR529 pKa = 11.84ISRR532 pKa = 11.84HH533 pKa = 3.54QCQKK537 pKa = 11.01KK538 pKa = 9.71KK539 pKa = 10.64LVNQGYY545 pKa = 10.15DD546 pKa = 3.43EE547 pKa = 4.64NLSEE551 pKa = 4.16YY552 pKa = 10.25QICTTVLKK560 pKa = 10.45YY561 pKa = 11.18NKK563 pKa = 9.77IFDD566 pKa = 4.04CGNFKK571 pKa = 9.11FTKK574 pKa = 10.39YY575 pKa = 10.77YY576 pKa = 9.77II577 pKa = 3.9
Molecular weight: 67.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
39404 |
343 |
3321 |
615.7 |
69.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.548 ± 0.212 | 1.178 ± 0.115 |
6.563 ± 0.219 | 5.779 ± 0.274 |
4.281 ± 0.107 | 6.357 ± 0.256 |
1.391 ± 0.088 | 7.588 ± 0.181 |
6.644 ± 0.294 | 7.301 ± 0.163 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.216 ± 0.096 | 7.606 ± 0.239 |
3.304 ± 0.11 | 3.672 ± 0.125 |
3.528 ± 0.128 | 8.382 ± 0.232 |
7.268 ± 0.266 | 6.228 ± 0.179 |
1.145 ± 0.057 | 5.017 ± 0.253 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
