
Paracoccus aminophilus JCM 7686
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus; Paracoccus aminophilus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
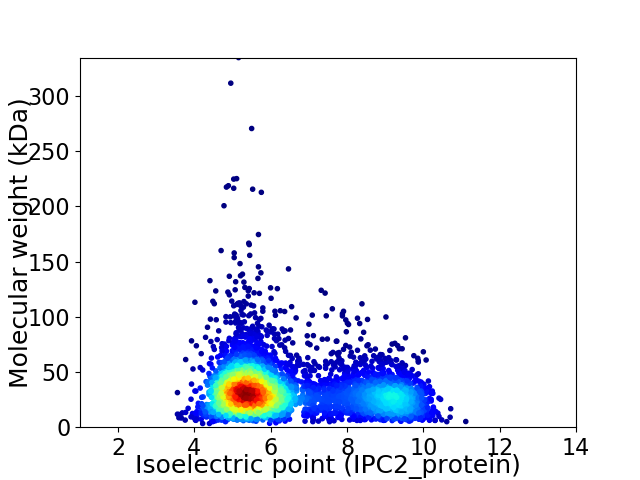
Virtual 2D-PAGE plot for 4462 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5XYM4|S5XYM4_PARAH ABC-type polar amino acid transport system ATPase component OS=Paracoccus aminophilus JCM 7686 OX=1367847 GN=JCM7686_1434 PE=3 SV=1
MM1 pKa = 7.07NQTLFLLLGVLGLAGLGSLMSSSSDD26 pKa = 3.04SHH28 pKa = 7.74GPEE31 pKa = 4.39TPPDD35 pKa = 3.93PYY37 pKa = 11.3GDD39 pKa = 3.49RR40 pKa = 11.84NVIEE44 pKa = 4.33GTEE47 pKa = 4.09GDD49 pKa = 3.95DD50 pKa = 4.32TIEE53 pKa = 4.26GTPGGDD59 pKa = 4.42AILTFDD65 pKa = 5.14GDD67 pKa = 3.89DD68 pKa = 3.65SVNGGGGDD76 pKa = 3.95DD77 pKa = 4.73IIWTGRR83 pKa = 11.84GNDD86 pKa = 3.79TVDD89 pKa = 3.19ATPGNDD95 pKa = 3.16LVYY98 pKa = 10.68LGEE101 pKa = 5.17GDD103 pKa = 3.9DD104 pKa = 5.39HH105 pKa = 7.99YY106 pKa = 11.66GAYY109 pKa = 10.38NPGVDD114 pKa = 3.86EE115 pKa = 4.54GHH117 pKa = 6.67DD118 pKa = 3.75TVAGGSGDD126 pKa = 4.52DD127 pKa = 4.14VIITNGGEE135 pKa = 3.54HH136 pKa = 7.13HH137 pKa = 7.32IYY139 pKa = 11.08GDD141 pKa = 4.16ADD143 pKa = 3.9PNDD146 pKa = 4.01DD147 pKa = 5.5SRR149 pKa = 11.84DD150 pKa = 3.64DD151 pKa = 3.78DD152 pKa = 4.06HH153 pKa = 9.26ASGNDD158 pKa = 3.48SIYY161 pKa = 11.15DD162 pKa = 3.52NGGTVYY168 pKa = 10.91VDD170 pKa = 3.82AGPGNDD176 pKa = 5.42LIWSPDD182 pKa = 3.72DD183 pKa = 3.8SDD185 pKa = 4.85KK186 pKa = 11.6DD187 pKa = 4.04HH188 pKa = 7.84PDD190 pKa = 3.08TLYY193 pKa = 11.3GGDD196 pKa = 4.11GDD198 pKa = 4.03DD199 pKa = 3.81TIYY202 pKa = 11.18AGGHH206 pKa = 6.91DD207 pKa = 4.28ILDD210 pKa = 4.02GGKK213 pKa = 10.74GSDD216 pKa = 3.6LYY218 pKa = 10.61IIRR221 pKa = 11.84SDD223 pKa = 3.04SSGPVDD229 pKa = 3.5ITYY232 pKa = 11.23GDD234 pKa = 3.91ADD236 pKa = 4.34NIQIALPADD245 pKa = 3.68YY246 pKa = 10.39TGDD249 pKa = 3.1KK250 pKa = 10.36HH251 pKa = 8.05FEE253 pKa = 4.03YY254 pKa = 10.99VQDD257 pKa = 3.71GDD259 pKa = 3.84NVRR262 pKa = 11.84LLVDD266 pKa = 4.11GNDD269 pKa = 3.35LALLRR274 pKa = 11.84DD275 pKa = 3.63VKK277 pKa = 10.63VANVRR282 pKa = 11.84DD283 pKa = 3.6VSFINQSDD291 pKa = 3.96APKK294 pKa = 10.72APWQTTT300 pKa = 3.13
MM1 pKa = 7.07NQTLFLLLGVLGLAGLGSLMSSSSDD26 pKa = 3.04SHH28 pKa = 7.74GPEE31 pKa = 4.39TPPDD35 pKa = 3.93PYY37 pKa = 11.3GDD39 pKa = 3.49RR40 pKa = 11.84NVIEE44 pKa = 4.33GTEE47 pKa = 4.09GDD49 pKa = 3.95DD50 pKa = 4.32TIEE53 pKa = 4.26GTPGGDD59 pKa = 4.42AILTFDD65 pKa = 5.14GDD67 pKa = 3.89DD68 pKa = 3.65SVNGGGGDD76 pKa = 3.95DD77 pKa = 4.73IIWTGRR83 pKa = 11.84GNDD86 pKa = 3.79TVDD89 pKa = 3.19ATPGNDD95 pKa = 3.16LVYY98 pKa = 10.68LGEE101 pKa = 5.17GDD103 pKa = 3.9DD104 pKa = 5.39HH105 pKa = 7.99YY106 pKa = 11.66GAYY109 pKa = 10.38NPGVDD114 pKa = 3.86EE115 pKa = 4.54GHH117 pKa = 6.67DD118 pKa = 3.75TVAGGSGDD126 pKa = 4.52DD127 pKa = 4.14VIITNGGEE135 pKa = 3.54HH136 pKa = 7.13HH137 pKa = 7.32IYY139 pKa = 11.08GDD141 pKa = 4.16ADD143 pKa = 3.9PNDD146 pKa = 4.01DD147 pKa = 5.5SRR149 pKa = 11.84DD150 pKa = 3.64DD151 pKa = 3.78DD152 pKa = 4.06HH153 pKa = 9.26ASGNDD158 pKa = 3.48SIYY161 pKa = 11.15DD162 pKa = 3.52NGGTVYY168 pKa = 10.91VDD170 pKa = 3.82AGPGNDD176 pKa = 5.42LIWSPDD182 pKa = 3.72DD183 pKa = 3.8SDD185 pKa = 4.85KK186 pKa = 11.6DD187 pKa = 4.04HH188 pKa = 7.84PDD190 pKa = 3.08TLYY193 pKa = 11.3GGDD196 pKa = 4.11GDD198 pKa = 4.03DD199 pKa = 3.81TIYY202 pKa = 11.18AGGHH206 pKa = 6.91DD207 pKa = 4.28ILDD210 pKa = 4.02GGKK213 pKa = 10.74GSDD216 pKa = 3.6LYY218 pKa = 10.61IIRR221 pKa = 11.84SDD223 pKa = 3.04SSGPVDD229 pKa = 3.5ITYY232 pKa = 11.23GDD234 pKa = 3.91ADD236 pKa = 4.34NIQIALPADD245 pKa = 3.68YY246 pKa = 10.39TGDD249 pKa = 3.1KK250 pKa = 10.36HH251 pKa = 8.05FEE253 pKa = 4.03YY254 pKa = 10.99VQDD257 pKa = 3.71GDD259 pKa = 3.84NVRR262 pKa = 11.84LLVDD266 pKa = 4.11GNDD269 pKa = 3.35LALLRR274 pKa = 11.84DD275 pKa = 3.63VKK277 pKa = 10.63VANVRR282 pKa = 11.84DD283 pKa = 3.6VSFINQSDD291 pKa = 3.96APKK294 pKa = 10.72APWQTTT300 pKa = 3.13
Molecular weight: 31.35 kDa
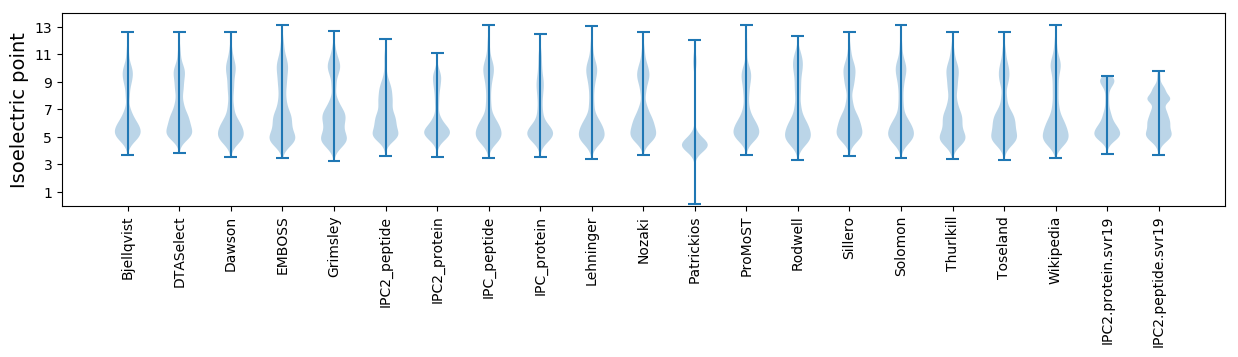
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5Y3I3|S5Y3I3_PARAH Selenide water dikinase OS=Paracoccus aminophilus JCM 7686 OX=1367847 GN=selD PE=3 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.2QPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.12GGRR29 pKa = 11.84RR30 pKa = 11.84VLNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.0GRR40 pKa = 11.84KK41 pKa = 8.91SLSAA45 pKa = 3.86
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.2QPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.12GGRR29 pKa = 11.84RR30 pKa = 11.84VLNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.0GRR40 pKa = 11.84KK41 pKa = 8.91SLSAA45 pKa = 3.86
Molecular weight: 5.16 kDa
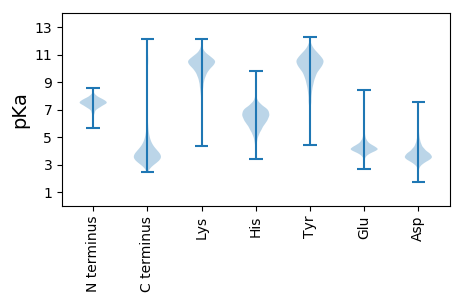
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1439745 |
30 |
3161 |
322.7 |
34.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.982 ± 0.051 | 0.794 ± 0.011 |
5.574 ± 0.029 | 5.743 ± 0.032 |
3.548 ± 0.022 | 8.732 ± 0.035 |
1.992 ± 0.016 | 5.273 ± 0.024 |
3.012 ± 0.032 | 10.551 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.527 ± 0.018 | 2.454 ± 0.022 |
5.435 ± 0.029 | 3.273 ± 0.021 |
7.138 ± 0.035 | 5.391 ± 0.027 |
5.261 ± 0.026 | 6.824 ± 0.029 |
1.416 ± 0.013 | 2.081 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
