
Beihai barnacle virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.81
Get precalculated fractions of proteins
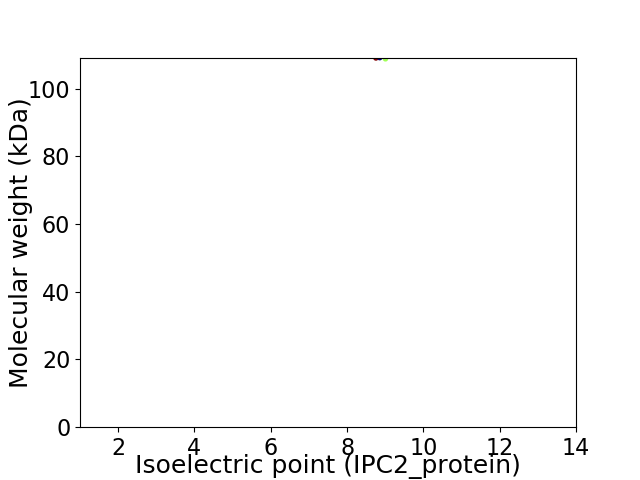
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI52|A0A1L3KI52_9VIRU RNA-dependent RNA polymerase OS=Beihai barnacle virus 10 OX=1922354 PE=4 SV=1
MM1 pKa = 7.5QSTNLLTSKK10 pKa = 10.48LIQTGCPRR18 pKa = 11.84APAGKK23 pKa = 9.79LGRR26 pKa = 11.84GYY28 pKa = 10.68RR29 pKa = 11.84IFIPHH34 pKa = 7.37LYY36 pKa = 10.6EE37 pKa = 5.34NGDD40 pKa = 3.51YY41 pKa = 11.07LLPGRR46 pKa = 11.84NHH48 pKa = 6.47SLLKK52 pKa = 10.64GVVVRR57 pKa = 11.84NRR59 pKa = 11.84MVKK62 pKa = 10.26PNLLISKK69 pKa = 7.68LTLMGFGKK77 pKa = 10.56RR78 pKa = 11.84FATLVSLRR86 pKa = 11.84PISHH90 pKa = 7.08IEE92 pKa = 3.72RR93 pKa = 11.84AEE95 pKa = 3.81QSVLGVIDD103 pKa = 4.49SLLLYY108 pKa = 10.53DD109 pKa = 4.46SEE111 pKa = 5.89LFITDD116 pKa = 3.27TGYY119 pKa = 11.76KK120 pKa = 9.54LLKK123 pKa = 10.45FIVKK127 pKa = 8.52KK128 pKa = 7.68TFKK131 pKa = 10.79VAIFNCALVTKK142 pKa = 9.64LWKK145 pKa = 10.37EE146 pKa = 3.37FSFYY150 pKa = 10.19VWAHH154 pKa = 3.91VTEE157 pKa = 4.55TVTRR161 pKa = 11.84EE162 pKa = 3.55KK163 pKa = 10.47PVRR166 pKa = 11.84KK167 pKa = 9.54RR168 pKa = 11.84EE169 pKa = 3.85NFFFSLVSEE178 pKa = 4.15QDD180 pKa = 3.14IEE182 pKa = 4.55VLKK185 pKa = 9.98LTLNKK190 pKa = 8.97KK191 pKa = 7.07TLTRR195 pKa = 11.84LAHH198 pKa = 5.65LTSSRR203 pKa = 11.84QFPPGEE209 pKa = 3.8RR210 pKa = 11.84QQEE213 pKa = 4.17IKK215 pKa = 10.49SLKK218 pKa = 10.3DD219 pKa = 3.36FEE221 pKa = 4.78SHH223 pKa = 4.46VTEE226 pKa = 4.87PYY228 pKa = 9.81IGNPVFLRR236 pKa = 11.84RR237 pKa = 11.84IRR239 pKa = 11.84LASRR243 pKa = 11.84VVGRR247 pKa = 11.84KK248 pKa = 8.7ARR250 pKa = 11.84RR251 pKa = 11.84LGMKK255 pKa = 9.86PLTDD259 pKa = 3.27SHH261 pKa = 7.22LSLAAAGSFYY271 pKa = 10.81TSVKK275 pKa = 10.47DD276 pKa = 3.41GGRR279 pKa = 11.84AEE281 pKa = 4.49EE282 pKa = 4.25LLNMLFKK289 pKa = 11.06YY290 pKa = 10.24LAYY293 pKa = 10.28VPKK296 pKa = 9.73EE297 pKa = 3.77SCEE300 pKa = 3.92IKK302 pKa = 10.11TPFYY306 pKa = 10.0TLKK309 pKa = 10.77DD310 pKa = 3.68VEE312 pKa = 4.76GVEE315 pKa = 5.31RR316 pKa = 11.84WRR318 pKa = 11.84TWCRR322 pKa = 11.84PAVYY326 pKa = 10.49DD327 pKa = 3.71HH328 pKa = 6.83FPNVSFGNLLPEE340 pKa = 4.05TLMGFEE346 pKa = 4.68TYY348 pKa = 10.52YY349 pKa = 10.72QGFDD353 pKa = 4.31EE354 pKa = 4.42ILGMQILACSYY365 pKa = 10.8LEE367 pKa = 3.99QEE369 pKa = 4.46EE370 pKa = 4.46HH371 pKa = 6.8LKK373 pKa = 10.9CNQNIPVRR381 pKa = 11.84VLTITEE387 pKa = 4.5PGSKK391 pKa = 10.25ARR393 pKa = 11.84IVTTGPCWLYY403 pKa = 10.24TLQQPVSHH411 pKa = 6.53VLRR414 pKa = 11.84GFLGQHH420 pKa = 6.55PSAAAGLTMSDD431 pKa = 3.75QAWQFLFLLEE441 pKa = 4.21KK442 pKa = 10.58AKK444 pKa = 10.87SHH446 pKa = 6.75FGDD449 pKa = 4.67DD450 pKa = 4.87FSCLSSDD457 pKa = 3.35LTAATDD463 pKa = 4.02VIPLCILKK471 pKa = 10.03EE472 pKa = 3.98LYY474 pKa = 10.41EE475 pKa = 4.48GFLEE479 pKa = 4.29GLRR482 pKa = 11.84IEE484 pKa = 5.27GPFLNIVGKK493 pKa = 9.46MIQLPRR499 pKa = 11.84LCSVEE504 pKa = 4.34KK505 pKa = 10.53INSYY509 pKa = 10.42FLNSRR514 pKa = 11.84GIFMGEE520 pKa = 4.28PIAKK524 pKa = 9.84VLLTLLNLSCEE535 pKa = 4.43EE536 pKa = 3.81IAIRR540 pKa = 11.84DD541 pKa = 3.71YY542 pKa = 11.58LGVDD546 pKa = 4.16FRR548 pKa = 11.84ASVQVRR554 pKa = 11.84WRR556 pKa = 11.84CFSVAGDD563 pKa = 3.45DD564 pKa = 4.55HH565 pKa = 7.63IAVGPVGYY573 pKa = 9.94LDD575 pKa = 5.56KK576 pKa = 10.6ITEE579 pKa = 4.17THH581 pKa = 6.06IKK583 pKa = 10.04AGSKK587 pKa = 10.13ISDD590 pKa = 3.76LKK592 pKa = 10.72HH593 pKa = 4.4GHH595 pKa = 5.29SRR597 pKa = 11.84IAVRR601 pKa = 11.84YY602 pKa = 8.23CEE604 pKa = 4.93KK605 pKa = 10.59ILDD608 pKa = 3.59VRR610 pKa = 11.84NFKK613 pKa = 10.91GSWSKK618 pKa = 9.18YY619 pKa = 7.59TINDD623 pKa = 3.16STEE626 pKa = 4.05AYY628 pKa = 9.56LASPFVDD635 pKa = 4.02SIKK638 pKa = 10.59VRR640 pKa = 11.84LLSPAAKK647 pKa = 9.98NVLSFNEE654 pKa = 4.01KK655 pKa = 8.53NTAFGKK661 pKa = 10.48GKK663 pKa = 10.43SLGRR667 pKa = 11.84TLQWLNKK674 pKa = 10.31DD675 pKa = 3.83CFDD678 pKa = 4.43SKK680 pKa = 10.44WISLVRR686 pKa = 11.84DD687 pKa = 3.33WFFKK691 pKa = 10.67RR692 pKa = 11.84MSSLLPDD699 pKa = 4.03RR700 pKa = 11.84SSGVYY705 pKa = 7.5WHH707 pKa = 7.08LLLPEE712 pKa = 4.27HH713 pKa = 6.82LGGLGLGTEE722 pKa = 4.71RR723 pKa = 11.84DD724 pKa = 3.85FEE726 pKa = 4.47DD727 pKa = 5.18LIVRR731 pKa = 11.84LPSPSRR737 pKa = 11.84TLLKK741 pKa = 10.58SIEE744 pKa = 4.5DD745 pKa = 3.74GNPNMGHH752 pKa = 6.26IRR754 pKa = 11.84LFSGFTSNTTYY765 pKa = 10.49RR766 pKa = 11.84GYY768 pKa = 10.82KK769 pKa = 9.45IPEE772 pKa = 4.09TEE774 pKa = 4.17KK775 pKa = 10.86EE776 pKa = 3.65IAALFFQGILEE787 pKa = 4.25EE788 pKa = 4.18EE789 pKa = 4.42KK790 pKa = 10.67SDD792 pKa = 4.11KK793 pKa = 11.02FEE795 pKa = 4.59NLVKK799 pKa = 10.64LYY801 pKa = 10.05RR802 pKa = 11.84YY803 pKa = 9.52EE804 pKa = 4.57DD805 pKa = 5.22LIPNTAIRR813 pKa = 11.84LLRR816 pKa = 11.84TQGWVRR822 pKa = 11.84KK823 pKa = 9.81DD824 pKa = 3.49YY825 pKa = 11.7LEE827 pKa = 4.9DD828 pKa = 4.71KK829 pKa = 10.33ILRR832 pKa = 11.84PFLFKK837 pKa = 10.74EE838 pKa = 3.9ILTGRR843 pKa = 11.84AKK845 pKa = 10.54VKK847 pKa = 10.73AFNTEE852 pKa = 4.05HH853 pKa = 6.64LKK855 pKa = 10.68SRR857 pKa = 11.84YY858 pKa = 9.12ARR860 pKa = 11.84LWDD863 pKa = 3.19LTYY866 pKa = 10.62RR867 pKa = 11.84GEE869 pKa = 4.23EE870 pKa = 4.19TISPQTLQVCFKK882 pKa = 9.79PPAYY886 pKa = 9.91HH887 pKa = 7.26LYY889 pKa = 10.59YY890 pKa = 11.2KK891 pKa = 10.14MGEE894 pKa = 4.06KK895 pKa = 10.61LEE897 pKa = 4.15LPFRR901 pKa = 11.84GRR903 pKa = 11.84VLKK906 pKa = 11.15VNLLEE911 pKa = 4.44EE912 pKa = 4.85GLLGMPDD919 pKa = 4.6LSIPWEE925 pKa = 4.21VIGDD929 pKa = 3.73ISGPLAPKK937 pKa = 10.02RR938 pKa = 11.84GNHH941 pKa = 5.12ARR943 pKa = 11.84SRR945 pKa = 11.84VIKK948 pKa = 10.2SPRR951 pKa = 11.84RR952 pKa = 11.84RR953 pKa = 11.84NSS955 pKa = 2.7
MM1 pKa = 7.5QSTNLLTSKK10 pKa = 10.48LIQTGCPRR18 pKa = 11.84APAGKK23 pKa = 9.79LGRR26 pKa = 11.84GYY28 pKa = 10.68RR29 pKa = 11.84IFIPHH34 pKa = 7.37LYY36 pKa = 10.6EE37 pKa = 5.34NGDD40 pKa = 3.51YY41 pKa = 11.07LLPGRR46 pKa = 11.84NHH48 pKa = 6.47SLLKK52 pKa = 10.64GVVVRR57 pKa = 11.84NRR59 pKa = 11.84MVKK62 pKa = 10.26PNLLISKK69 pKa = 7.68LTLMGFGKK77 pKa = 10.56RR78 pKa = 11.84FATLVSLRR86 pKa = 11.84PISHH90 pKa = 7.08IEE92 pKa = 3.72RR93 pKa = 11.84AEE95 pKa = 3.81QSVLGVIDD103 pKa = 4.49SLLLYY108 pKa = 10.53DD109 pKa = 4.46SEE111 pKa = 5.89LFITDD116 pKa = 3.27TGYY119 pKa = 11.76KK120 pKa = 9.54LLKK123 pKa = 10.45FIVKK127 pKa = 8.52KK128 pKa = 7.68TFKK131 pKa = 10.79VAIFNCALVTKK142 pKa = 9.64LWKK145 pKa = 10.37EE146 pKa = 3.37FSFYY150 pKa = 10.19VWAHH154 pKa = 3.91VTEE157 pKa = 4.55TVTRR161 pKa = 11.84EE162 pKa = 3.55KK163 pKa = 10.47PVRR166 pKa = 11.84KK167 pKa = 9.54RR168 pKa = 11.84EE169 pKa = 3.85NFFFSLVSEE178 pKa = 4.15QDD180 pKa = 3.14IEE182 pKa = 4.55VLKK185 pKa = 9.98LTLNKK190 pKa = 8.97KK191 pKa = 7.07TLTRR195 pKa = 11.84LAHH198 pKa = 5.65LTSSRR203 pKa = 11.84QFPPGEE209 pKa = 3.8RR210 pKa = 11.84QQEE213 pKa = 4.17IKK215 pKa = 10.49SLKK218 pKa = 10.3DD219 pKa = 3.36FEE221 pKa = 4.78SHH223 pKa = 4.46VTEE226 pKa = 4.87PYY228 pKa = 9.81IGNPVFLRR236 pKa = 11.84RR237 pKa = 11.84IRR239 pKa = 11.84LASRR243 pKa = 11.84VVGRR247 pKa = 11.84KK248 pKa = 8.7ARR250 pKa = 11.84RR251 pKa = 11.84LGMKK255 pKa = 9.86PLTDD259 pKa = 3.27SHH261 pKa = 7.22LSLAAAGSFYY271 pKa = 10.81TSVKK275 pKa = 10.47DD276 pKa = 3.41GGRR279 pKa = 11.84AEE281 pKa = 4.49EE282 pKa = 4.25LLNMLFKK289 pKa = 11.06YY290 pKa = 10.24LAYY293 pKa = 10.28VPKK296 pKa = 9.73EE297 pKa = 3.77SCEE300 pKa = 3.92IKK302 pKa = 10.11TPFYY306 pKa = 10.0TLKK309 pKa = 10.77DD310 pKa = 3.68VEE312 pKa = 4.76GVEE315 pKa = 5.31RR316 pKa = 11.84WRR318 pKa = 11.84TWCRR322 pKa = 11.84PAVYY326 pKa = 10.49DD327 pKa = 3.71HH328 pKa = 6.83FPNVSFGNLLPEE340 pKa = 4.05TLMGFEE346 pKa = 4.68TYY348 pKa = 10.52YY349 pKa = 10.72QGFDD353 pKa = 4.31EE354 pKa = 4.42ILGMQILACSYY365 pKa = 10.8LEE367 pKa = 3.99QEE369 pKa = 4.46EE370 pKa = 4.46HH371 pKa = 6.8LKK373 pKa = 10.9CNQNIPVRR381 pKa = 11.84VLTITEE387 pKa = 4.5PGSKK391 pKa = 10.25ARR393 pKa = 11.84IVTTGPCWLYY403 pKa = 10.24TLQQPVSHH411 pKa = 6.53VLRR414 pKa = 11.84GFLGQHH420 pKa = 6.55PSAAAGLTMSDD431 pKa = 3.75QAWQFLFLLEE441 pKa = 4.21KK442 pKa = 10.58AKK444 pKa = 10.87SHH446 pKa = 6.75FGDD449 pKa = 4.67DD450 pKa = 4.87FSCLSSDD457 pKa = 3.35LTAATDD463 pKa = 4.02VIPLCILKK471 pKa = 10.03EE472 pKa = 3.98LYY474 pKa = 10.41EE475 pKa = 4.48GFLEE479 pKa = 4.29GLRR482 pKa = 11.84IEE484 pKa = 5.27GPFLNIVGKK493 pKa = 9.46MIQLPRR499 pKa = 11.84LCSVEE504 pKa = 4.34KK505 pKa = 10.53INSYY509 pKa = 10.42FLNSRR514 pKa = 11.84GIFMGEE520 pKa = 4.28PIAKK524 pKa = 9.84VLLTLLNLSCEE535 pKa = 4.43EE536 pKa = 3.81IAIRR540 pKa = 11.84DD541 pKa = 3.71YY542 pKa = 11.58LGVDD546 pKa = 4.16FRR548 pKa = 11.84ASVQVRR554 pKa = 11.84WRR556 pKa = 11.84CFSVAGDD563 pKa = 3.45DD564 pKa = 4.55HH565 pKa = 7.63IAVGPVGYY573 pKa = 9.94LDD575 pKa = 5.56KK576 pKa = 10.6ITEE579 pKa = 4.17THH581 pKa = 6.06IKK583 pKa = 10.04AGSKK587 pKa = 10.13ISDD590 pKa = 3.76LKK592 pKa = 10.72HH593 pKa = 4.4GHH595 pKa = 5.29SRR597 pKa = 11.84IAVRR601 pKa = 11.84YY602 pKa = 8.23CEE604 pKa = 4.93KK605 pKa = 10.59ILDD608 pKa = 3.59VRR610 pKa = 11.84NFKK613 pKa = 10.91GSWSKK618 pKa = 9.18YY619 pKa = 7.59TINDD623 pKa = 3.16STEE626 pKa = 4.05AYY628 pKa = 9.56LASPFVDD635 pKa = 4.02SIKK638 pKa = 10.59VRR640 pKa = 11.84LLSPAAKK647 pKa = 9.98NVLSFNEE654 pKa = 4.01KK655 pKa = 8.53NTAFGKK661 pKa = 10.48GKK663 pKa = 10.43SLGRR667 pKa = 11.84TLQWLNKK674 pKa = 10.31DD675 pKa = 3.83CFDD678 pKa = 4.43SKK680 pKa = 10.44WISLVRR686 pKa = 11.84DD687 pKa = 3.33WFFKK691 pKa = 10.67RR692 pKa = 11.84MSSLLPDD699 pKa = 4.03RR700 pKa = 11.84SSGVYY705 pKa = 7.5WHH707 pKa = 7.08LLLPEE712 pKa = 4.27HH713 pKa = 6.82LGGLGLGTEE722 pKa = 4.71RR723 pKa = 11.84DD724 pKa = 3.85FEE726 pKa = 4.47DD727 pKa = 5.18LIVRR731 pKa = 11.84LPSPSRR737 pKa = 11.84TLLKK741 pKa = 10.58SIEE744 pKa = 4.5DD745 pKa = 3.74GNPNMGHH752 pKa = 6.26IRR754 pKa = 11.84LFSGFTSNTTYY765 pKa = 10.49RR766 pKa = 11.84GYY768 pKa = 10.82KK769 pKa = 9.45IPEE772 pKa = 4.09TEE774 pKa = 4.17KK775 pKa = 10.86EE776 pKa = 3.65IAALFFQGILEE787 pKa = 4.25EE788 pKa = 4.18EE789 pKa = 4.42KK790 pKa = 10.67SDD792 pKa = 4.11KK793 pKa = 11.02FEE795 pKa = 4.59NLVKK799 pKa = 10.64LYY801 pKa = 10.05RR802 pKa = 11.84YY803 pKa = 9.52EE804 pKa = 4.57DD805 pKa = 5.22LIPNTAIRR813 pKa = 11.84LLRR816 pKa = 11.84TQGWVRR822 pKa = 11.84KK823 pKa = 9.81DD824 pKa = 3.49YY825 pKa = 11.7LEE827 pKa = 4.9DD828 pKa = 4.71KK829 pKa = 10.33ILRR832 pKa = 11.84PFLFKK837 pKa = 10.74EE838 pKa = 3.9ILTGRR843 pKa = 11.84AKK845 pKa = 10.54VKK847 pKa = 10.73AFNTEE852 pKa = 4.05HH853 pKa = 6.64LKK855 pKa = 10.68SRR857 pKa = 11.84YY858 pKa = 9.12ARR860 pKa = 11.84LWDD863 pKa = 3.19LTYY866 pKa = 10.62RR867 pKa = 11.84GEE869 pKa = 4.23EE870 pKa = 4.19TISPQTLQVCFKK882 pKa = 9.79PPAYY886 pKa = 9.91HH887 pKa = 7.26LYY889 pKa = 10.59YY890 pKa = 11.2KK891 pKa = 10.14MGEE894 pKa = 4.06KK895 pKa = 10.61LEE897 pKa = 4.15LPFRR901 pKa = 11.84GRR903 pKa = 11.84VLKK906 pKa = 11.15VNLLEE911 pKa = 4.44EE912 pKa = 4.85GLLGMPDD919 pKa = 4.6LSIPWEE925 pKa = 4.21VIGDD929 pKa = 3.73ISGPLAPKK937 pKa = 10.02RR938 pKa = 11.84GNHH941 pKa = 5.12ARR943 pKa = 11.84SRR945 pKa = 11.84VIKK948 pKa = 10.2SPRR951 pKa = 11.84RR952 pKa = 11.84RR953 pKa = 11.84NSS955 pKa = 2.7
Molecular weight: 109.03 kDa
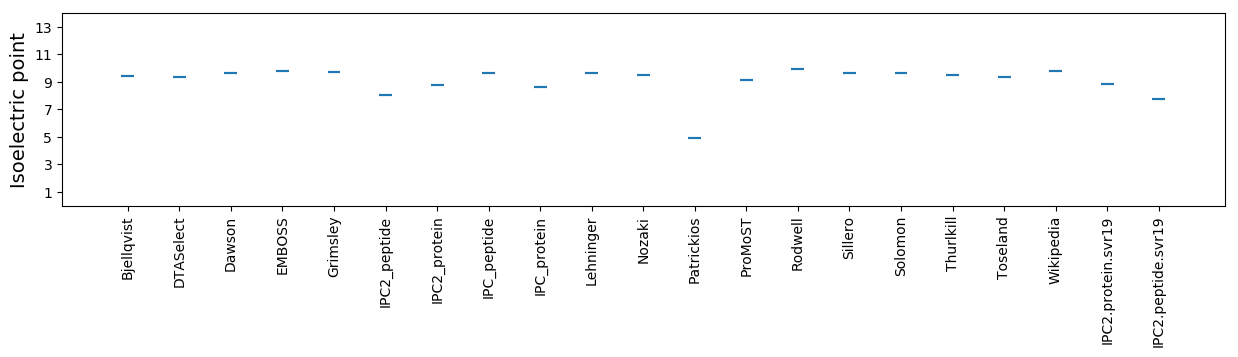
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI52|A0A1L3KI52_9VIRU RNA-dependent RNA polymerase OS=Beihai barnacle virus 10 OX=1922354 PE=4 SV=1
MM1 pKa = 7.5QSTNLLTSKK10 pKa = 10.48LIQTGCPRR18 pKa = 11.84APAGKK23 pKa = 9.79LGRR26 pKa = 11.84GYY28 pKa = 10.68RR29 pKa = 11.84IFIPHH34 pKa = 7.37LYY36 pKa = 10.6EE37 pKa = 5.34NGDD40 pKa = 3.51YY41 pKa = 11.07LLPGRR46 pKa = 11.84NHH48 pKa = 6.47SLLKK52 pKa = 10.64GVVVRR57 pKa = 11.84NRR59 pKa = 11.84MVKK62 pKa = 10.26PNLLISKK69 pKa = 7.68LTLMGFGKK77 pKa = 10.56RR78 pKa = 11.84FATLVSLRR86 pKa = 11.84PISHH90 pKa = 7.08IEE92 pKa = 3.72RR93 pKa = 11.84AEE95 pKa = 3.81QSVLGVIDD103 pKa = 4.49SLLLYY108 pKa = 10.53DD109 pKa = 4.46SEE111 pKa = 5.89LFITDD116 pKa = 3.27TGYY119 pKa = 11.76KK120 pKa = 9.54LLKK123 pKa = 10.45FIVKK127 pKa = 8.52KK128 pKa = 7.68TFKK131 pKa = 10.79VAIFNCALVTKK142 pKa = 9.64LWKK145 pKa = 10.37EE146 pKa = 3.37FSFYY150 pKa = 10.19VWAHH154 pKa = 3.91VTEE157 pKa = 4.55TVTRR161 pKa = 11.84EE162 pKa = 3.55KK163 pKa = 10.47PVRR166 pKa = 11.84KK167 pKa = 9.54RR168 pKa = 11.84EE169 pKa = 3.85NFFFSLVSEE178 pKa = 4.15QDD180 pKa = 3.14IEE182 pKa = 4.55VLKK185 pKa = 9.98LTLNKK190 pKa = 8.97KK191 pKa = 7.07TLTRR195 pKa = 11.84LAHH198 pKa = 5.65LTSSRR203 pKa = 11.84QFPPGEE209 pKa = 3.8RR210 pKa = 11.84QQEE213 pKa = 4.17IKK215 pKa = 10.49SLKK218 pKa = 10.3DD219 pKa = 3.36FEE221 pKa = 4.78SHH223 pKa = 4.46VTEE226 pKa = 4.87PYY228 pKa = 9.81IGNPVFLRR236 pKa = 11.84RR237 pKa = 11.84IRR239 pKa = 11.84LASRR243 pKa = 11.84VVGRR247 pKa = 11.84KK248 pKa = 8.7ARR250 pKa = 11.84RR251 pKa = 11.84LGMKK255 pKa = 9.86PLTDD259 pKa = 3.27SHH261 pKa = 7.22LSLAAAGSFYY271 pKa = 10.81TSVKK275 pKa = 10.47DD276 pKa = 3.41GGRR279 pKa = 11.84AEE281 pKa = 4.49EE282 pKa = 4.25LLNMLFKK289 pKa = 11.06YY290 pKa = 10.24LAYY293 pKa = 10.28VPKK296 pKa = 9.73EE297 pKa = 3.77SCEE300 pKa = 3.92IKK302 pKa = 10.11TPFYY306 pKa = 10.0TLKK309 pKa = 10.77DD310 pKa = 3.68VEE312 pKa = 4.76GVEE315 pKa = 5.31RR316 pKa = 11.84WRR318 pKa = 11.84TWCRR322 pKa = 11.84PAVYY326 pKa = 10.49DD327 pKa = 3.71HH328 pKa = 6.83FPNVSFGNLLPEE340 pKa = 4.05TLMGFEE346 pKa = 4.68TYY348 pKa = 10.52YY349 pKa = 10.72QGFDD353 pKa = 4.31EE354 pKa = 4.42ILGMQILACSYY365 pKa = 10.8LEE367 pKa = 3.99QEE369 pKa = 4.46EE370 pKa = 4.46HH371 pKa = 6.8LKK373 pKa = 10.9CNQNIPVRR381 pKa = 11.84VLTITEE387 pKa = 4.5PGSKK391 pKa = 10.25ARR393 pKa = 11.84IVTTGPCWLYY403 pKa = 10.24TLQQPVSHH411 pKa = 6.53VLRR414 pKa = 11.84GFLGQHH420 pKa = 6.55PSAAAGLTMSDD431 pKa = 3.75QAWQFLFLLEE441 pKa = 4.21KK442 pKa = 10.58AKK444 pKa = 10.87SHH446 pKa = 6.75FGDD449 pKa = 4.67DD450 pKa = 4.87FSCLSSDD457 pKa = 3.35LTAATDD463 pKa = 4.02VIPLCILKK471 pKa = 10.03EE472 pKa = 3.98LYY474 pKa = 10.41EE475 pKa = 4.48GFLEE479 pKa = 4.29GLRR482 pKa = 11.84IEE484 pKa = 5.27GPFLNIVGKK493 pKa = 9.46MIQLPRR499 pKa = 11.84LCSVEE504 pKa = 4.34KK505 pKa = 10.53INSYY509 pKa = 10.42FLNSRR514 pKa = 11.84GIFMGEE520 pKa = 4.28PIAKK524 pKa = 9.84VLLTLLNLSCEE535 pKa = 4.43EE536 pKa = 3.81IAIRR540 pKa = 11.84DD541 pKa = 3.71YY542 pKa = 11.58LGVDD546 pKa = 4.16FRR548 pKa = 11.84ASVQVRR554 pKa = 11.84WRR556 pKa = 11.84CFSVAGDD563 pKa = 3.45DD564 pKa = 4.55HH565 pKa = 7.63IAVGPVGYY573 pKa = 9.94LDD575 pKa = 5.56KK576 pKa = 10.6ITEE579 pKa = 4.17THH581 pKa = 6.06IKK583 pKa = 10.04AGSKK587 pKa = 10.13ISDD590 pKa = 3.76LKK592 pKa = 10.72HH593 pKa = 4.4GHH595 pKa = 5.29SRR597 pKa = 11.84IAVRR601 pKa = 11.84YY602 pKa = 8.23CEE604 pKa = 4.93KK605 pKa = 10.59ILDD608 pKa = 3.59VRR610 pKa = 11.84NFKK613 pKa = 10.91GSWSKK618 pKa = 9.18YY619 pKa = 7.59TINDD623 pKa = 3.16STEE626 pKa = 4.05AYY628 pKa = 9.56LASPFVDD635 pKa = 4.02SIKK638 pKa = 10.59VRR640 pKa = 11.84LLSPAAKK647 pKa = 9.98NVLSFNEE654 pKa = 4.01KK655 pKa = 8.53NTAFGKK661 pKa = 10.48GKK663 pKa = 10.43SLGRR667 pKa = 11.84TLQWLNKK674 pKa = 10.31DD675 pKa = 3.83CFDD678 pKa = 4.43SKK680 pKa = 10.44WISLVRR686 pKa = 11.84DD687 pKa = 3.33WFFKK691 pKa = 10.67RR692 pKa = 11.84MSSLLPDD699 pKa = 4.03RR700 pKa = 11.84SSGVYY705 pKa = 7.5WHH707 pKa = 7.08LLLPEE712 pKa = 4.27HH713 pKa = 6.82LGGLGLGTEE722 pKa = 4.71RR723 pKa = 11.84DD724 pKa = 3.85FEE726 pKa = 4.47DD727 pKa = 5.18LIVRR731 pKa = 11.84LPSPSRR737 pKa = 11.84TLLKK741 pKa = 10.58SIEE744 pKa = 4.5DD745 pKa = 3.74GNPNMGHH752 pKa = 6.26IRR754 pKa = 11.84LFSGFTSNTTYY765 pKa = 10.49RR766 pKa = 11.84GYY768 pKa = 10.82KK769 pKa = 9.45IPEE772 pKa = 4.09TEE774 pKa = 4.17KK775 pKa = 10.86EE776 pKa = 3.65IAALFFQGILEE787 pKa = 4.25EE788 pKa = 4.18EE789 pKa = 4.42KK790 pKa = 10.67SDD792 pKa = 4.11KK793 pKa = 11.02FEE795 pKa = 4.59NLVKK799 pKa = 10.64LYY801 pKa = 10.05RR802 pKa = 11.84YY803 pKa = 9.52EE804 pKa = 4.57DD805 pKa = 5.22LIPNTAIRR813 pKa = 11.84LLRR816 pKa = 11.84TQGWVRR822 pKa = 11.84KK823 pKa = 9.81DD824 pKa = 3.49YY825 pKa = 11.7LEE827 pKa = 4.9DD828 pKa = 4.71KK829 pKa = 10.33ILRR832 pKa = 11.84PFLFKK837 pKa = 10.74EE838 pKa = 3.9ILTGRR843 pKa = 11.84AKK845 pKa = 10.54VKK847 pKa = 10.73AFNTEE852 pKa = 4.05HH853 pKa = 6.64LKK855 pKa = 10.68SRR857 pKa = 11.84YY858 pKa = 9.12ARR860 pKa = 11.84LWDD863 pKa = 3.19LTYY866 pKa = 10.62RR867 pKa = 11.84GEE869 pKa = 4.23EE870 pKa = 4.19TISPQTLQVCFKK882 pKa = 9.79PPAYY886 pKa = 9.91HH887 pKa = 7.26LYY889 pKa = 10.59YY890 pKa = 11.2KK891 pKa = 10.14MGEE894 pKa = 4.06KK895 pKa = 10.61LEE897 pKa = 4.15LPFRR901 pKa = 11.84GRR903 pKa = 11.84VLKK906 pKa = 11.15VNLLEE911 pKa = 4.44EE912 pKa = 4.85GLLGMPDD919 pKa = 4.6LSIPWEE925 pKa = 4.21VIGDD929 pKa = 3.73ISGPLAPKK937 pKa = 10.02RR938 pKa = 11.84GNHH941 pKa = 5.12ARR943 pKa = 11.84SRR945 pKa = 11.84VIKK948 pKa = 10.2SPRR951 pKa = 11.84RR952 pKa = 11.84RR953 pKa = 11.84NSS955 pKa = 2.7
MM1 pKa = 7.5QSTNLLTSKK10 pKa = 10.48LIQTGCPRR18 pKa = 11.84APAGKK23 pKa = 9.79LGRR26 pKa = 11.84GYY28 pKa = 10.68RR29 pKa = 11.84IFIPHH34 pKa = 7.37LYY36 pKa = 10.6EE37 pKa = 5.34NGDD40 pKa = 3.51YY41 pKa = 11.07LLPGRR46 pKa = 11.84NHH48 pKa = 6.47SLLKK52 pKa = 10.64GVVVRR57 pKa = 11.84NRR59 pKa = 11.84MVKK62 pKa = 10.26PNLLISKK69 pKa = 7.68LTLMGFGKK77 pKa = 10.56RR78 pKa = 11.84FATLVSLRR86 pKa = 11.84PISHH90 pKa = 7.08IEE92 pKa = 3.72RR93 pKa = 11.84AEE95 pKa = 3.81QSVLGVIDD103 pKa = 4.49SLLLYY108 pKa = 10.53DD109 pKa = 4.46SEE111 pKa = 5.89LFITDD116 pKa = 3.27TGYY119 pKa = 11.76KK120 pKa = 9.54LLKK123 pKa = 10.45FIVKK127 pKa = 8.52KK128 pKa = 7.68TFKK131 pKa = 10.79VAIFNCALVTKK142 pKa = 9.64LWKK145 pKa = 10.37EE146 pKa = 3.37FSFYY150 pKa = 10.19VWAHH154 pKa = 3.91VTEE157 pKa = 4.55TVTRR161 pKa = 11.84EE162 pKa = 3.55KK163 pKa = 10.47PVRR166 pKa = 11.84KK167 pKa = 9.54RR168 pKa = 11.84EE169 pKa = 3.85NFFFSLVSEE178 pKa = 4.15QDD180 pKa = 3.14IEE182 pKa = 4.55VLKK185 pKa = 9.98LTLNKK190 pKa = 8.97KK191 pKa = 7.07TLTRR195 pKa = 11.84LAHH198 pKa = 5.65LTSSRR203 pKa = 11.84QFPPGEE209 pKa = 3.8RR210 pKa = 11.84QQEE213 pKa = 4.17IKK215 pKa = 10.49SLKK218 pKa = 10.3DD219 pKa = 3.36FEE221 pKa = 4.78SHH223 pKa = 4.46VTEE226 pKa = 4.87PYY228 pKa = 9.81IGNPVFLRR236 pKa = 11.84RR237 pKa = 11.84IRR239 pKa = 11.84LASRR243 pKa = 11.84VVGRR247 pKa = 11.84KK248 pKa = 8.7ARR250 pKa = 11.84RR251 pKa = 11.84LGMKK255 pKa = 9.86PLTDD259 pKa = 3.27SHH261 pKa = 7.22LSLAAAGSFYY271 pKa = 10.81TSVKK275 pKa = 10.47DD276 pKa = 3.41GGRR279 pKa = 11.84AEE281 pKa = 4.49EE282 pKa = 4.25LLNMLFKK289 pKa = 11.06YY290 pKa = 10.24LAYY293 pKa = 10.28VPKK296 pKa = 9.73EE297 pKa = 3.77SCEE300 pKa = 3.92IKK302 pKa = 10.11TPFYY306 pKa = 10.0TLKK309 pKa = 10.77DD310 pKa = 3.68VEE312 pKa = 4.76GVEE315 pKa = 5.31RR316 pKa = 11.84WRR318 pKa = 11.84TWCRR322 pKa = 11.84PAVYY326 pKa = 10.49DD327 pKa = 3.71HH328 pKa = 6.83FPNVSFGNLLPEE340 pKa = 4.05TLMGFEE346 pKa = 4.68TYY348 pKa = 10.52YY349 pKa = 10.72QGFDD353 pKa = 4.31EE354 pKa = 4.42ILGMQILACSYY365 pKa = 10.8LEE367 pKa = 3.99QEE369 pKa = 4.46EE370 pKa = 4.46HH371 pKa = 6.8LKK373 pKa = 10.9CNQNIPVRR381 pKa = 11.84VLTITEE387 pKa = 4.5PGSKK391 pKa = 10.25ARR393 pKa = 11.84IVTTGPCWLYY403 pKa = 10.24TLQQPVSHH411 pKa = 6.53VLRR414 pKa = 11.84GFLGQHH420 pKa = 6.55PSAAAGLTMSDD431 pKa = 3.75QAWQFLFLLEE441 pKa = 4.21KK442 pKa = 10.58AKK444 pKa = 10.87SHH446 pKa = 6.75FGDD449 pKa = 4.67DD450 pKa = 4.87FSCLSSDD457 pKa = 3.35LTAATDD463 pKa = 4.02VIPLCILKK471 pKa = 10.03EE472 pKa = 3.98LYY474 pKa = 10.41EE475 pKa = 4.48GFLEE479 pKa = 4.29GLRR482 pKa = 11.84IEE484 pKa = 5.27GPFLNIVGKK493 pKa = 9.46MIQLPRR499 pKa = 11.84LCSVEE504 pKa = 4.34KK505 pKa = 10.53INSYY509 pKa = 10.42FLNSRR514 pKa = 11.84GIFMGEE520 pKa = 4.28PIAKK524 pKa = 9.84VLLTLLNLSCEE535 pKa = 4.43EE536 pKa = 3.81IAIRR540 pKa = 11.84DD541 pKa = 3.71YY542 pKa = 11.58LGVDD546 pKa = 4.16FRR548 pKa = 11.84ASVQVRR554 pKa = 11.84WRR556 pKa = 11.84CFSVAGDD563 pKa = 3.45DD564 pKa = 4.55HH565 pKa = 7.63IAVGPVGYY573 pKa = 9.94LDD575 pKa = 5.56KK576 pKa = 10.6ITEE579 pKa = 4.17THH581 pKa = 6.06IKK583 pKa = 10.04AGSKK587 pKa = 10.13ISDD590 pKa = 3.76LKK592 pKa = 10.72HH593 pKa = 4.4GHH595 pKa = 5.29SRR597 pKa = 11.84IAVRR601 pKa = 11.84YY602 pKa = 8.23CEE604 pKa = 4.93KK605 pKa = 10.59ILDD608 pKa = 3.59VRR610 pKa = 11.84NFKK613 pKa = 10.91GSWSKK618 pKa = 9.18YY619 pKa = 7.59TINDD623 pKa = 3.16STEE626 pKa = 4.05AYY628 pKa = 9.56LASPFVDD635 pKa = 4.02SIKK638 pKa = 10.59VRR640 pKa = 11.84LLSPAAKK647 pKa = 9.98NVLSFNEE654 pKa = 4.01KK655 pKa = 8.53NTAFGKK661 pKa = 10.48GKK663 pKa = 10.43SLGRR667 pKa = 11.84TLQWLNKK674 pKa = 10.31DD675 pKa = 3.83CFDD678 pKa = 4.43SKK680 pKa = 10.44WISLVRR686 pKa = 11.84DD687 pKa = 3.33WFFKK691 pKa = 10.67RR692 pKa = 11.84MSSLLPDD699 pKa = 4.03RR700 pKa = 11.84SSGVYY705 pKa = 7.5WHH707 pKa = 7.08LLLPEE712 pKa = 4.27HH713 pKa = 6.82LGGLGLGTEE722 pKa = 4.71RR723 pKa = 11.84DD724 pKa = 3.85FEE726 pKa = 4.47DD727 pKa = 5.18LIVRR731 pKa = 11.84LPSPSRR737 pKa = 11.84TLLKK741 pKa = 10.58SIEE744 pKa = 4.5DD745 pKa = 3.74GNPNMGHH752 pKa = 6.26IRR754 pKa = 11.84LFSGFTSNTTYY765 pKa = 10.49RR766 pKa = 11.84GYY768 pKa = 10.82KK769 pKa = 9.45IPEE772 pKa = 4.09TEE774 pKa = 4.17KK775 pKa = 10.86EE776 pKa = 3.65IAALFFQGILEE787 pKa = 4.25EE788 pKa = 4.18EE789 pKa = 4.42KK790 pKa = 10.67SDD792 pKa = 4.11KK793 pKa = 11.02FEE795 pKa = 4.59NLVKK799 pKa = 10.64LYY801 pKa = 10.05RR802 pKa = 11.84YY803 pKa = 9.52EE804 pKa = 4.57DD805 pKa = 5.22LIPNTAIRR813 pKa = 11.84LLRR816 pKa = 11.84TQGWVRR822 pKa = 11.84KK823 pKa = 9.81DD824 pKa = 3.49YY825 pKa = 11.7LEE827 pKa = 4.9DD828 pKa = 4.71KK829 pKa = 10.33ILRR832 pKa = 11.84PFLFKK837 pKa = 10.74EE838 pKa = 3.9ILTGRR843 pKa = 11.84AKK845 pKa = 10.54VKK847 pKa = 10.73AFNTEE852 pKa = 4.05HH853 pKa = 6.64LKK855 pKa = 10.68SRR857 pKa = 11.84YY858 pKa = 9.12ARR860 pKa = 11.84LWDD863 pKa = 3.19LTYY866 pKa = 10.62RR867 pKa = 11.84GEE869 pKa = 4.23EE870 pKa = 4.19TISPQTLQVCFKK882 pKa = 9.79PPAYY886 pKa = 9.91HH887 pKa = 7.26LYY889 pKa = 10.59YY890 pKa = 11.2KK891 pKa = 10.14MGEE894 pKa = 4.06KK895 pKa = 10.61LEE897 pKa = 4.15LPFRR901 pKa = 11.84GRR903 pKa = 11.84VLKK906 pKa = 11.15VNLLEE911 pKa = 4.44EE912 pKa = 4.85GLLGMPDD919 pKa = 4.6LSIPWEE925 pKa = 4.21VIGDD929 pKa = 3.73ISGPLAPKK937 pKa = 10.02RR938 pKa = 11.84GNHH941 pKa = 5.12ARR943 pKa = 11.84SRR945 pKa = 11.84VIKK948 pKa = 10.2SPRR951 pKa = 11.84RR952 pKa = 11.84RR953 pKa = 11.84NSS955 pKa = 2.7
Molecular weight: 109.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
955 |
955 |
955 |
955.0 |
109.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.817 ± 0.0 | 1.571 ± 0.0 |
4.084 ± 0.0 | 6.492 ± 0.0 |
5.34 ± 0.0 | 6.806 ± 0.0 |
2.304 ± 0.0 | 5.654 ± 0.0 |
7.12 ± 0.0 | 13.089 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.466 ± 0.0 | 3.455 ± 0.0 |
4.817 ± 0.0 | 2.408 ± 0.0 |
6.911 ± 0.0 | 7.12 ± 0.0 |
5.445 ± 0.0 | 5.969 ± 0.0 |
1.571 ± 0.0 | 3.56 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
