
Curvularia thermal tolerance virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Curvulaviridae; Orthocurvulavirus; Curvularia orthocurvulavirus 1
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
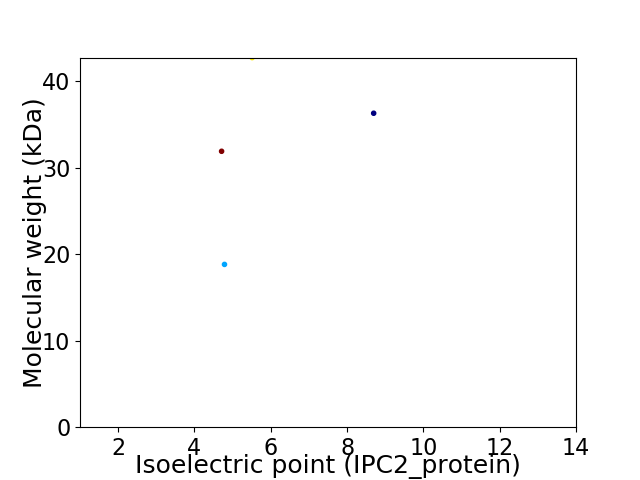
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
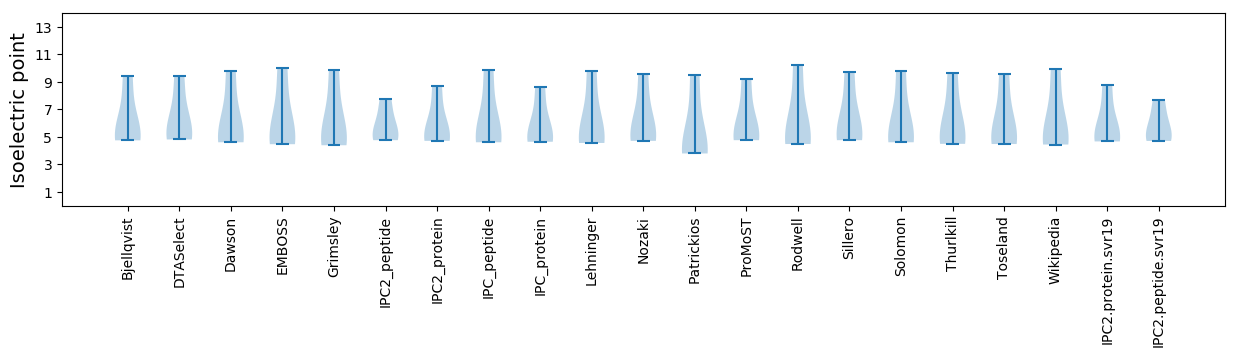
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3EYA9|A3EYA9_9VIRU Uncharacterized protein OS=Curvularia thermal tolerance virus OX=421976 PE=4 SV=1
MM1 pKa = 7.56AAMSAMTRR9 pKa = 11.84TGTLFANLSWRR20 pKa = 11.84LLSAGNDD27 pKa = 4.07GIRR30 pKa = 11.84LQKK33 pKa = 10.54RR34 pKa = 11.84VGTTSGHH41 pKa = 4.66SHH43 pKa = 5.52NTLLQSICTLIVGYY57 pKa = 9.81GVLMATHH64 pKa = 7.72PDD66 pKa = 3.43LTDD69 pKa = 3.52EE70 pKa = 4.81EE71 pKa = 4.5IKK73 pKa = 10.77AAAQIEE79 pKa = 4.61SLGDD83 pKa = 3.66DD84 pKa = 4.36NITGTKK90 pKa = 10.35APLQPLPVEE99 pKa = 4.54DD100 pKa = 4.35VADD103 pKa = 4.14KK104 pKa = 10.76AWDD107 pKa = 3.4MFGIDD112 pKa = 3.27WSGKK116 pKa = 9.74KK117 pKa = 10.14SFATTAVLDD126 pKa = 3.72ATALQFQGVQYY137 pKa = 10.03LGKK140 pKa = 8.68YY141 pKa = 8.16FRR143 pKa = 11.84RR144 pKa = 11.84EE145 pKa = 3.96EE146 pKa = 4.19YY147 pKa = 10.1PLDD150 pKa = 3.85EE151 pKa = 4.49GTVVVAIPYY160 pKa = 10.06RR161 pKa = 11.84PFKK164 pKa = 9.56EE165 pKa = 4.21TYY167 pKa = 10.02LRR169 pKa = 11.84LLYY172 pKa = 9.93PEE174 pKa = 4.58YY175 pKa = 11.08GSLEE179 pKa = 4.07ADD181 pKa = 3.66QTWLRR186 pKa = 11.84LLGNYY191 pKa = 9.51LDD193 pKa = 4.5AAGNPVTEE201 pKa = 4.21KK202 pKa = 9.99WLQGFMDD209 pKa = 3.77WLEE212 pKa = 4.26PQVVAPPQVWPSNFQRR228 pKa = 11.84MVSRR232 pKa = 11.84DD233 pKa = 3.32YY234 pKa = 11.49SGIGIEE240 pKa = 4.14MPRR243 pKa = 11.84PEE245 pKa = 4.29RR246 pKa = 11.84MCYY249 pKa = 7.86EE250 pKa = 3.72QWRR253 pKa = 11.84DD254 pKa = 3.54LVVLPRR260 pKa = 11.84DD261 pKa = 4.68DD262 pKa = 5.58YY263 pKa = 10.67RR264 pKa = 11.84QLWKK268 pKa = 10.88ASDD271 pKa = 4.06PNDD274 pKa = 3.73DD275 pKa = 3.92ALYY278 pKa = 8.89EE279 pKa = 4.03AEE281 pKa = 4.31YY282 pKa = 11.25
MM1 pKa = 7.56AAMSAMTRR9 pKa = 11.84TGTLFANLSWRR20 pKa = 11.84LLSAGNDD27 pKa = 4.07GIRR30 pKa = 11.84LQKK33 pKa = 10.54RR34 pKa = 11.84VGTTSGHH41 pKa = 4.66SHH43 pKa = 5.52NTLLQSICTLIVGYY57 pKa = 9.81GVLMATHH64 pKa = 7.72PDD66 pKa = 3.43LTDD69 pKa = 3.52EE70 pKa = 4.81EE71 pKa = 4.5IKK73 pKa = 10.77AAAQIEE79 pKa = 4.61SLGDD83 pKa = 3.66DD84 pKa = 4.36NITGTKK90 pKa = 10.35APLQPLPVEE99 pKa = 4.54DD100 pKa = 4.35VADD103 pKa = 4.14KK104 pKa = 10.76AWDD107 pKa = 3.4MFGIDD112 pKa = 3.27WSGKK116 pKa = 9.74KK117 pKa = 10.14SFATTAVLDD126 pKa = 3.72ATALQFQGVQYY137 pKa = 10.03LGKK140 pKa = 8.68YY141 pKa = 8.16FRR143 pKa = 11.84RR144 pKa = 11.84EE145 pKa = 3.96EE146 pKa = 4.19YY147 pKa = 10.1PLDD150 pKa = 3.85EE151 pKa = 4.49GTVVVAIPYY160 pKa = 10.06RR161 pKa = 11.84PFKK164 pKa = 9.56EE165 pKa = 4.21TYY167 pKa = 10.02LRR169 pKa = 11.84LLYY172 pKa = 9.93PEE174 pKa = 4.58YY175 pKa = 11.08GSLEE179 pKa = 4.07ADD181 pKa = 3.66QTWLRR186 pKa = 11.84LLGNYY191 pKa = 9.51LDD193 pKa = 4.5AAGNPVTEE201 pKa = 4.21KK202 pKa = 9.99WLQGFMDD209 pKa = 3.77WLEE212 pKa = 4.26PQVVAPPQVWPSNFQRR228 pKa = 11.84MVSRR232 pKa = 11.84DD233 pKa = 3.32YY234 pKa = 11.49SGIGIEE240 pKa = 4.14MPRR243 pKa = 11.84PEE245 pKa = 4.29RR246 pKa = 11.84MCYY249 pKa = 7.86EE250 pKa = 3.72QWRR253 pKa = 11.84DD254 pKa = 3.54LVVLPRR260 pKa = 11.84DD261 pKa = 4.68DD262 pKa = 5.58YY263 pKa = 10.67RR264 pKa = 11.84QLWKK268 pKa = 10.88ASDD271 pKa = 4.06PNDD274 pKa = 3.73DD275 pKa = 3.92ALYY278 pKa = 8.89EE279 pKa = 4.03AEE281 pKa = 4.31YY282 pKa = 11.25
Molecular weight: 31.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3EYB0|A3EYB0_9VIRU Uncharacterized protein OS=Curvularia thermal tolerance virus OX=421976 PE=4 SV=1
MM1 pKa = 8.14SGMKK5 pKa = 10.37APVSFLEE12 pKa = 4.39KK13 pKa = 10.84YY14 pKa = 10.55GGATANVDD22 pKa = 4.1FSEE25 pKa = 5.27GPSLPAEE32 pKa = 4.37VLPMPEE38 pKa = 4.58PTDD41 pKa = 3.7SLQTLSNKK49 pKa = 8.33MRR51 pKa = 11.84ALAEE55 pKa = 4.11VVEE58 pKa = 5.12RR59 pKa = 11.84GASVARR65 pKa = 11.84FTGMLGTPQGSRR77 pKa = 11.84DD78 pKa = 3.53AEE80 pKa = 4.11EE81 pKa = 4.03VGKK84 pKa = 10.63LRR86 pKa = 11.84MTDD89 pKa = 3.63LEE91 pKa = 4.65RR92 pKa = 11.84EE93 pKa = 4.39MFDD96 pKa = 2.56HH97 pKa = 6.79WAAGGVLPPIDD108 pKa = 3.82WDD110 pKa = 3.54KK111 pKa = 11.96DD112 pKa = 3.49MKK114 pKa = 10.48KK115 pKa = 10.39APSSGEE121 pKa = 3.66GMRR124 pKa = 11.84RR125 pKa = 11.84AQRR128 pKa = 11.84RR129 pKa = 11.84AQALNMLYY137 pKa = 8.51KK138 pKa = 9.79TDD140 pKa = 3.66KK141 pKa = 10.78AQPQHH146 pKa = 6.06VMWYY150 pKa = 8.04QDD152 pKa = 3.13TYY154 pKa = 11.0ATIIPLLVAVTKK166 pKa = 10.79VIGAQTDD173 pKa = 3.9FLSGQDD179 pKa = 3.37KK180 pKa = 9.55LTDD183 pKa = 3.51NEE185 pKa = 4.13VAEE188 pKa = 4.32VQTIGRR194 pKa = 11.84ATGTTYY200 pKa = 10.4ATINAIQEE208 pKa = 4.31EE209 pKa = 4.86VHH211 pKa = 6.92KK212 pKa = 10.93LLANINRR219 pKa = 11.84SKK221 pKa = 11.16DD222 pKa = 3.3VLQARR227 pKa = 11.84EE228 pKa = 3.86HH229 pKa = 6.57AIKK232 pKa = 10.6AKK234 pKa = 10.19NPEE237 pKa = 4.2YY238 pKa = 10.24KK239 pKa = 10.22KK240 pKa = 10.71KK241 pKa = 10.52RR242 pKa = 11.84SPEE245 pKa = 3.49NDD247 pKa = 2.77RR248 pKa = 11.84RR249 pKa = 11.84GLLGKK254 pKa = 10.2RR255 pKa = 11.84KK256 pKa = 9.73IGVEE260 pKa = 3.84VDD262 pKa = 4.33FGQAPEE268 pKa = 3.82PSRR271 pKa = 11.84RR272 pKa = 11.84ARR274 pKa = 11.84RR275 pKa = 11.84DD276 pKa = 2.9LAQGIRR282 pKa = 11.84PRR284 pKa = 11.84SVIEE288 pKa = 3.77AAKK291 pKa = 8.99ATAEE295 pKa = 4.01SRR297 pKa = 11.84EE298 pKa = 4.26KK299 pKa = 10.83GKK301 pKa = 10.35QPAKK305 pKa = 9.77EE306 pKa = 4.23GNGTPPTAGRR316 pKa = 11.84RR317 pKa = 11.84DD318 pKa = 3.21GHH320 pKa = 5.83FQLGGWQDD328 pKa = 3.3TGVV331 pKa = 3.14
MM1 pKa = 8.14SGMKK5 pKa = 10.37APVSFLEE12 pKa = 4.39KK13 pKa = 10.84YY14 pKa = 10.55GGATANVDD22 pKa = 4.1FSEE25 pKa = 5.27GPSLPAEE32 pKa = 4.37VLPMPEE38 pKa = 4.58PTDD41 pKa = 3.7SLQTLSNKK49 pKa = 8.33MRR51 pKa = 11.84ALAEE55 pKa = 4.11VVEE58 pKa = 5.12RR59 pKa = 11.84GASVARR65 pKa = 11.84FTGMLGTPQGSRR77 pKa = 11.84DD78 pKa = 3.53AEE80 pKa = 4.11EE81 pKa = 4.03VGKK84 pKa = 10.63LRR86 pKa = 11.84MTDD89 pKa = 3.63LEE91 pKa = 4.65RR92 pKa = 11.84EE93 pKa = 4.39MFDD96 pKa = 2.56HH97 pKa = 6.79WAAGGVLPPIDD108 pKa = 3.82WDD110 pKa = 3.54KK111 pKa = 11.96DD112 pKa = 3.49MKK114 pKa = 10.48KK115 pKa = 10.39APSSGEE121 pKa = 3.66GMRR124 pKa = 11.84RR125 pKa = 11.84AQRR128 pKa = 11.84RR129 pKa = 11.84AQALNMLYY137 pKa = 8.51KK138 pKa = 9.79TDD140 pKa = 3.66KK141 pKa = 10.78AQPQHH146 pKa = 6.06VMWYY150 pKa = 8.04QDD152 pKa = 3.13TYY154 pKa = 11.0ATIIPLLVAVTKK166 pKa = 10.79VIGAQTDD173 pKa = 3.9FLSGQDD179 pKa = 3.37KK180 pKa = 9.55LTDD183 pKa = 3.51NEE185 pKa = 4.13VAEE188 pKa = 4.32VQTIGRR194 pKa = 11.84ATGTTYY200 pKa = 10.4ATINAIQEE208 pKa = 4.31EE209 pKa = 4.86VHH211 pKa = 6.92KK212 pKa = 10.93LLANINRR219 pKa = 11.84SKK221 pKa = 11.16DD222 pKa = 3.3VLQARR227 pKa = 11.84EE228 pKa = 3.86HH229 pKa = 6.57AIKK232 pKa = 10.6AKK234 pKa = 10.19NPEE237 pKa = 4.2YY238 pKa = 10.24KK239 pKa = 10.22KK240 pKa = 10.71KK241 pKa = 10.52RR242 pKa = 11.84SPEE245 pKa = 3.49NDD247 pKa = 2.77RR248 pKa = 11.84RR249 pKa = 11.84GLLGKK254 pKa = 10.2RR255 pKa = 11.84KK256 pKa = 9.73IGVEE260 pKa = 3.84VDD262 pKa = 4.33FGQAPEE268 pKa = 3.82PSRR271 pKa = 11.84RR272 pKa = 11.84ARR274 pKa = 11.84RR275 pKa = 11.84DD276 pKa = 2.9LAQGIRR282 pKa = 11.84PRR284 pKa = 11.84SVIEE288 pKa = 3.77AAKK291 pKa = 8.99ATAEE295 pKa = 4.01SRR297 pKa = 11.84EE298 pKa = 4.26KK299 pKa = 10.83GKK301 pKa = 10.35QPAKK305 pKa = 9.77EE306 pKa = 4.23GNGTPPTAGRR316 pKa = 11.84RR317 pKa = 11.84DD318 pKa = 3.21GHH320 pKa = 5.83FQLGGWQDD328 pKa = 3.3TGVV331 pKa = 3.14
Molecular weight: 36.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1152 |
168 |
371 |
288.0 |
32.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.028 ± 0.689 | 0.521 ± 0.185 |
6.337 ± 0.534 | 7.726 ± 0.712 |
3.125 ± 0.518 | 7.118 ± 1.086 |
1.563 ± 0.171 | 3.472 ± 0.103 |
4.774 ± 0.926 | 9.375 ± 0.924 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.125 ± 0.089 | 2.778 ± 0.109 |
5.903 ± 0.232 | 4.253 ± 0.562 |
7.378 ± 0.529 | 5.729 ± 0.878 |
6.25 ± 0.18 | 5.816 ± 0.172 |
2.344 ± 0.513 | 3.385 ± 0.576 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
