
Cedecea neteri
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Cedecea
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
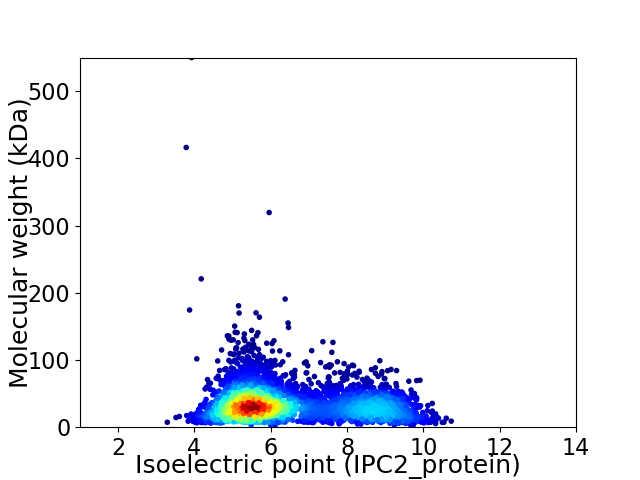
Virtual 2D-PAGE plot for 4293 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A089PZ75|A0A089PZ75_9ENTR Acetylornithine/succinyldiaminopimelate aminotransferase OS=Cedecea neteri OX=158822 GN=argD PE=3 SV=1
MM1 pKa = 7.35TKK3 pKa = 10.42VLLCVGNSMMGDD15 pKa = 3.65DD16 pKa = 4.5GAGPLLAEE24 pKa = 4.76KK25 pKa = 10.38CQASPQGDD33 pKa = 3.32WVVIDD38 pKa = 4.49GGSAPEE44 pKa = 3.93NDD46 pKa = 2.96VVAIRR51 pKa = 11.84EE52 pKa = 4.17LHH54 pKa = 6.59PEE56 pKa = 3.82LLLIVDD62 pKa = 4.5ATDD65 pKa = 3.01MMLNPGEE72 pKa = 4.29IRR74 pKa = 11.84VIDD77 pKa = 4.32PDD79 pKa = 5.18DD80 pKa = 3.61IAEE83 pKa = 4.13MFMMTTHH90 pKa = 6.45NMPLNYY96 pKa = 10.23LVDD99 pKa = 3.78QLQEE103 pKa = 4.11DD104 pKa = 3.84VGEE107 pKa = 4.36VVFLGIQPDD116 pKa = 3.43IVGFYY121 pKa = 11.02YY122 pKa = 10.43PMTEE126 pKa = 3.73AVKK129 pKa = 10.37AAVEE133 pKa = 3.92QVYY136 pKa = 10.01QRR138 pKa = 11.84LATWSGYY145 pKa = 10.35GGYY148 pKa = 10.15AQLTAEE154 pKa = 4.34EE155 pKa = 4.79TEE157 pKa = 4.43GLL159 pKa = 3.87
MM1 pKa = 7.35TKK3 pKa = 10.42VLLCVGNSMMGDD15 pKa = 3.65DD16 pKa = 4.5GAGPLLAEE24 pKa = 4.76KK25 pKa = 10.38CQASPQGDD33 pKa = 3.32WVVIDD38 pKa = 4.49GGSAPEE44 pKa = 3.93NDD46 pKa = 2.96VVAIRR51 pKa = 11.84EE52 pKa = 4.17LHH54 pKa = 6.59PEE56 pKa = 3.82LLLIVDD62 pKa = 4.5ATDD65 pKa = 3.01MMLNPGEE72 pKa = 4.29IRR74 pKa = 11.84VIDD77 pKa = 4.32PDD79 pKa = 5.18DD80 pKa = 3.61IAEE83 pKa = 4.13MFMMTTHH90 pKa = 6.45NMPLNYY96 pKa = 10.23LVDD99 pKa = 3.78QLQEE103 pKa = 4.11DD104 pKa = 3.84VGEE107 pKa = 4.36VVFLGIQPDD116 pKa = 3.43IVGFYY121 pKa = 11.02YY122 pKa = 10.43PMTEE126 pKa = 3.73AVKK129 pKa = 10.37AAVEE133 pKa = 3.92QVYY136 pKa = 10.01QRR138 pKa = 11.84LATWSGYY145 pKa = 10.35GGYY148 pKa = 10.15AQLTAEE154 pKa = 4.34EE155 pKa = 4.79TEE157 pKa = 4.43GLL159 pKa = 3.87
Molecular weight: 17.34 kDa
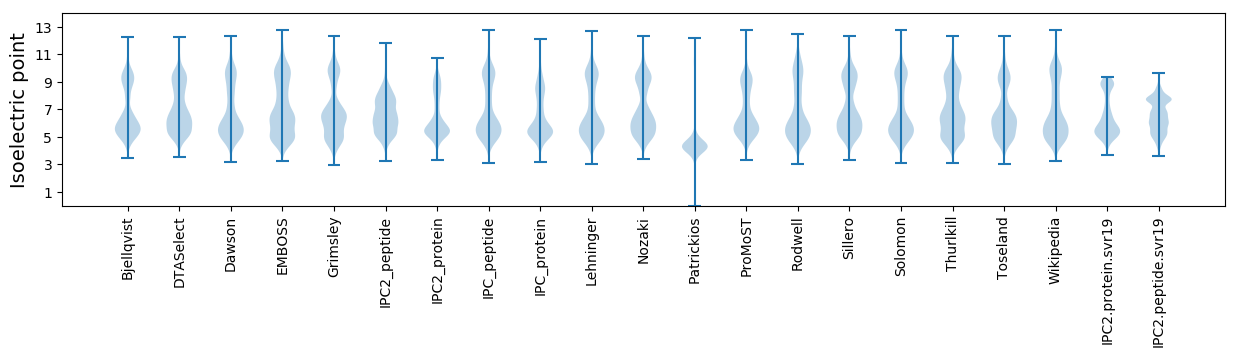
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A089PYJ0|A0A089PYJ0_9ENTR Protein PsiE homolog OS=Cedecea neteri OX=158822 GN=psiE PE=3 SV=1
MM1 pKa = 7.58PALAGGAVRR10 pKa = 11.84IILLLLCAFTLFLTGYY26 pKa = 9.53NFQQMLPAGLWWQAITLPQVTDD48 pKa = 3.2VSQMLFHH55 pKa = 6.66YY56 pKa = 10.31SLLPRR61 pKa = 11.84TTLALLTGAGLALAGCLFQHH81 pKa = 7.43ILRR84 pKa = 11.84NPLAEE89 pKa = 4.27PATLGVAAGAQLGLTLATLFLAGAGEE115 pKa = 4.39TGRR118 pKa = 11.84QLAALAGAMVVGSIVLGAAWGKK140 pKa = 10.44RR141 pKa = 11.84MSPVTLILAGLVLGLYY157 pKa = 10.06CGAVKK162 pKa = 10.66SFLVLFNHH170 pKa = 6.53EE171 pKa = 4.11RR172 pKa = 11.84LQNLFIWSSGMLNQYY187 pKa = 9.95DD188 pKa = 3.55WAGVEE193 pKa = 4.29FLWPRR198 pKa = 11.84LLAGLVLIVLMIRR211 pKa = 11.84PLGMLALDD219 pKa = 3.74DD220 pKa = 3.93TVLRR224 pKa = 11.84GLGMKK229 pKa = 10.15LALVRR234 pKa = 11.84VGGLFLALLLSSMLVSVVGVIGFIGLFAPVLAGMFGVRR272 pKa = 11.84RR273 pKa = 11.84LLPKK277 pKa = 10.45LLASMATGALLLLLSDD293 pKa = 4.9QLVIWLGSHH302 pKa = 4.52WQEE305 pKa = 4.22LPTGAVTALVGAPLMLWLLPRR326 pKa = 11.84LRR328 pKa = 11.84HH329 pKa = 5.25QRR331 pKa = 11.84LAATQDD337 pKa = 2.97ASIAAEE343 pKa = 3.88RR344 pKa = 11.84HH345 pKa = 5.33LSPGTVLLMCAALLLVALLALGVGRR370 pKa = 11.84EE371 pKa = 3.98SAGWFVGLQEE381 pKa = 3.95MWQWRR386 pKa = 11.84WPRR389 pKa = 11.84VLSAIAAGAMLAAAGTLVQKK409 pKa = 7.85MTGNPMASPEE419 pKa = 4.03VLGVSSGAACAIVLLTFLVPGDD441 pKa = 3.46VSAWQLPAGFAGATLTLLAMLVLARR466 pKa = 11.84TGLAPGRR473 pKa = 11.84LLLTGVALSTVFATLLTLYY492 pKa = 9.9LASGDD497 pKa = 3.91PRR499 pKa = 11.84SGAVLTWLSGSTWKK513 pKa = 8.71VTGQQALVSGGMALVLIGLTPLVRR537 pKa = 11.84RR538 pKa = 11.84WLTILPLGGEE548 pKa = 4.3AAQALGVALSRR559 pKa = 11.84SRR561 pKa = 11.84ISILVLAAGLVAAATLCVGPLSFVGLMAPHH591 pKa = 7.08MARR594 pKa = 11.84MLGFRR599 pKa = 11.84RR600 pKa = 11.84AVPQLVVAIALGAGLMLVADD620 pKa = 3.91WLGRR624 pKa = 11.84SILFPFQIPAGLLATLCGAPYY645 pKa = 9.81FIWLLRR651 pKa = 11.84KK652 pKa = 9.43AA653 pKa = 4.13
MM1 pKa = 7.58PALAGGAVRR10 pKa = 11.84IILLLLCAFTLFLTGYY26 pKa = 9.53NFQQMLPAGLWWQAITLPQVTDD48 pKa = 3.2VSQMLFHH55 pKa = 6.66YY56 pKa = 10.31SLLPRR61 pKa = 11.84TTLALLTGAGLALAGCLFQHH81 pKa = 7.43ILRR84 pKa = 11.84NPLAEE89 pKa = 4.27PATLGVAAGAQLGLTLATLFLAGAGEE115 pKa = 4.39TGRR118 pKa = 11.84QLAALAGAMVVGSIVLGAAWGKK140 pKa = 10.44RR141 pKa = 11.84MSPVTLILAGLVLGLYY157 pKa = 10.06CGAVKK162 pKa = 10.66SFLVLFNHH170 pKa = 6.53EE171 pKa = 4.11RR172 pKa = 11.84LQNLFIWSSGMLNQYY187 pKa = 9.95DD188 pKa = 3.55WAGVEE193 pKa = 4.29FLWPRR198 pKa = 11.84LLAGLVLIVLMIRR211 pKa = 11.84PLGMLALDD219 pKa = 3.74DD220 pKa = 3.93TVLRR224 pKa = 11.84GLGMKK229 pKa = 10.15LALVRR234 pKa = 11.84VGGLFLALLLSSMLVSVVGVIGFIGLFAPVLAGMFGVRR272 pKa = 11.84RR273 pKa = 11.84LLPKK277 pKa = 10.45LLASMATGALLLLLSDD293 pKa = 4.9QLVIWLGSHH302 pKa = 4.52WQEE305 pKa = 4.22LPTGAVTALVGAPLMLWLLPRR326 pKa = 11.84LRR328 pKa = 11.84HH329 pKa = 5.25QRR331 pKa = 11.84LAATQDD337 pKa = 2.97ASIAAEE343 pKa = 3.88RR344 pKa = 11.84HH345 pKa = 5.33LSPGTVLLMCAALLLVALLALGVGRR370 pKa = 11.84EE371 pKa = 3.98SAGWFVGLQEE381 pKa = 3.95MWQWRR386 pKa = 11.84WPRR389 pKa = 11.84VLSAIAAGAMLAAAGTLVQKK409 pKa = 7.85MTGNPMASPEE419 pKa = 4.03VLGVSSGAACAIVLLTFLVPGDD441 pKa = 3.46VSAWQLPAGFAGATLTLLAMLVLARR466 pKa = 11.84TGLAPGRR473 pKa = 11.84LLLTGVALSTVFATLLTLYY492 pKa = 9.9LASGDD497 pKa = 3.91PRR499 pKa = 11.84SGAVLTWLSGSTWKK513 pKa = 8.71VTGQQALVSGGMALVLIGLTPLVRR537 pKa = 11.84RR538 pKa = 11.84WLTILPLGGEE548 pKa = 4.3AAQALGVALSRR559 pKa = 11.84SRR561 pKa = 11.84ISILVLAAGLVAAATLCVGPLSFVGLMAPHH591 pKa = 7.08MARR594 pKa = 11.84MLGFRR599 pKa = 11.84RR600 pKa = 11.84AVPQLVVAIALGAGLMLVADD620 pKa = 3.91WLGRR624 pKa = 11.84SILFPFQIPAGLLATLCGAPYY645 pKa = 9.81FIWLLRR651 pKa = 11.84KK652 pKa = 9.43AA653 pKa = 4.13
Molecular weight: 68.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1387837 |
29 |
5414 |
323.3 |
35.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.912 ± 0.042 | 0.993 ± 0.015 |
5.118 ± 0.024 | 5.683 ± 0.041 |
3.833 ± 0.023 | 7.625 ± 0.046 |
2.196 ± 0.019 | 5.629 ± 0.031 |
4.227 ± 0.031 | 11.026 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.719 ± 0.02 | 3.714 ± 0.028 |
4.483 ± 0.026 | 4.501 ± 0.032 |
5.559 ± 0.041 | 6.07 ± 0.035 |
5.304 ± 0.049 | 7.209 ± 0.034 |
1.505 ± 0.016 | 2.694 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
