
Scheffersomyces segobiensis virus L
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Totivirus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
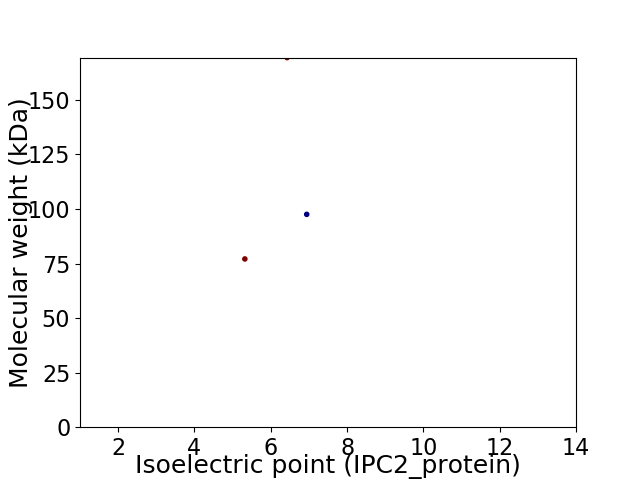
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4MEM7|M4MEM7_9VIRU Isoform of M4MG99 RNA-directed RNA polymerase (Fragment) OS=Scheffersomyces segobiensis virus L OX=1300323 GN=pol PE=3 SV=1
MM1 pKa = 7.85SYY3 pKa = 10.93QLFDD7 pKa = 3.66QLTGPLANVLPNFKK21 pKa = 10.55YY22 pKa = 9.93LAKK25 pKa = 9.13TVKK28 pKa = 10.32YY29 pKa = 10.04GIKK32 pKa = 10.05ADD34 pKa = 3.41VDD36 pKa = 3.31IGVTSEE42 pKa = 3.62FTNAYY47 pKa = 8.5KK48 pKa = 10.55RR49 pKa = 11.84RR50 pKa = 11.84GALFTNVLSPFGVFNAEE67 pKa = 4.27VVRR70 pKa = 11.84DD71 pKa = 3.8PPIFDD76 pKa = 5.18GINARR81 pKa = 11.84FFNDD85 pKa = 2.91YY86 pKa = 11.56GEE88 pKa = 4.14IDD90 pKa = 3.41EE91 pKa = 4.6RR92 pKa = 11.84AIAEE96 pKa = 4.23TLRR99 pKa = 11.84LKK101 pKa = 9.56GTYY104 pKa = 9.06RR105 pKa = 11.84PNVIAEE111 pKa = 4.65LAPTIASCNSEE122 pKa = 3.75NHH124 pKa = 5.34VSVLVNLLRR133 pKa = 11.84FSIIKK138 pKa = 10.46RR139 pKa = 11.84SDD141 pKa = 3.1PKK143 pKa = 11.03DD144 pKa = 3.64FKK146 pKa = 11.07FVNDD150 pKa = 4.16DD151 pKa = 3.41LFYY154 pKa = 11.14DD155 pKa = 4.35DD156 pKa = 4.02GHH158 pKa = 7.31LAVSYY163 pKa = 11.0KK164 pKa = 10.84NYY166 pKa = 10.59LKK168 pKa = 10.82RR169 pKa = 11.84EE170 pKa = 4.09FNEE173 pKa = 3.52EE174 pKa = 3.6LVFIFRR180 pKa = 11.84EE181 pKa = 5.32GIQIEE186 pKa = 4.4RR187 pKa = 11.84FSEE190 pKa = 4.3STVVGDD196 pKa = 6.22DD197 pKa = 4.42FYY199 pKa = 11.75CPNIEE204 pKa = 4.02NLSARR209 pKa = 11.84EE210 pKa = 3.84FNILLTLLGSIKK222 pKa = 10.14CDD224 pKa = 3.37YY225 pKa = 10.06PLRR228 pKa = 11.84LAFSTPRR235 pKa = 11.84LVDD238 pKa = 3.54KK239 pKa = 11.01LVLPLTTKK247 pKa = 9.58HH248 pKa = 5.54ASYY251 pKa = 11.2YY252 pKa = 10.38EE253 pKa = 4.05VVNDD257 pKa = 3.82FTANEE262 pKa = 3.73VDD264 pKa = 3.68QIIRR268 pKa = 11.84KK269 pKa = 9.48YY270 pKa = 9.76IFANRR275 pKa = 11.84VEE277 pKa = 4.39TAFDD281 pKa = 3.65LAYY284 pKa = 10.49LIVVTAMFAPLPRR297 pKa = 11.84AIEE300 pKa = 4.1ANGWVSPVNLIKK312 pKa = 10.77LPNVGSVRR320 pKa = 11.84GLMPEE325 pKa = 3.91MTGGQPLSKK334 pKa = 10.32SPQKK338 pKa = 11.1ALTWLNYY345 pKa = 8.51KK346 pKa = 10.07QSPGRR351 pKa = 11.84IAIHH355 pKa = 7.13AIAACEE361 pKa = 3.6AYY363 pKa = 8.82YY364 pKa = 10.45TGFFEE369 pKa = 5.9VLTASPNGIEE379 pKa = 4.42DD380 pKa = 3.98TLTAIGVSSSVTATPYY396 pKa = 11.59KK397 pKa = 9.74MFCEE401 pKa = 4.08SAAYY405 pKa = 10.19RR406 pKa = 11.84FGKK409 pKa = 10.35EE410 pKa = 4.46FDD412 pKa = 5.32LIWLTNAGVDD422 pKa = 4.14CYY424 pKa = 11.54SHH426 pKa = 7.26LLATEE431 pKa = 4.39PAALSILAKK440 pKa = 10.72VVDD443 pKa = 4.39TEE445 pKa = 4.4LEE447 pKa = 4.4GYY449 pKa = 9.94DD450 pKa = 3.96VYY452 pKa = 9.28TTSAFGEE459 pKa = 4.46DD460 pKa = 3.59TVHH463 pKa = 7.31VITKK467 pKa = 7.57EE468 pKa = 3.95VKK470 pKa = 8.6PALFPVLSMGINDD483 pKa = 4.24DD484 pKa = 4.18RR485 pKa = 11.84YY486 pKa = 9.19FANSLEE492 pKa = 4.24YY493 pKa = 10.7SATLYY498 pKa = 10.83ADD500 pKa = 3.19HH501 pKa = 7.12SEE503 pKa = 4.28GVLATTEE510 pKa = 3.99SDD512 pKa = 3.68HH513 pKa = 6.18MNKK516 pKa = 9.98ALSILRR522 pKa = 11.84IGGYY526 pKa = 9.84DD527 pKa = 3.0AVMTDD532 pKa = 3.38RR533 pKa = 11.84LSGLSYY539 pKa = 10.92RR540 pKa = 11.84NWAANSNGQVLPMLPPKK557 pKa = 10.67LKK559 pKa = 10.67GKK561 pKa = 7.74STYY564 pKa = 10.36VIPFNTIVKK573 pKa = 10.06RR574 pKa = 11.84KK575 pKa = 8.63KK576 pKa = 9.55SWMEE580 pKa = 3.76LGSLTDD586 pKa = 3.83SVTMTAKK593 pKa = 10.26INLRR597 pKa = 11.84GYY599 pKa = 11.24VIMTNGKK606 pKa = 9.09LVSTYY611 pKa = 10.29LPKK614 pKa = 10.47YY615 pKa = 7.52QTINVYY621 pKa = 10.03PKK623 pKa = 10.42SLNKK627 pKa = 10.36EE628 pKa = 3.7EE629 pKa = 4.11MVAVEE634 pKa = 4.74MKK636 pKa = 10.44TSATTIKK643 pKa = 10.23YY644 pKa = 9.64RR645 pKa = 11.84YY646 pKa = 9.88SGFLLAASQVEE657 pKa = 4.3APPARR662 pKa = 11.84LQLLSTDD669 pKa = 3.59ILGSQPIEE677 pKa = 4.16TSSEE681 pKa = 3.7EE682 pKa = 3.93AGAVGDD688 pKa = 4.16DD689 pKa = 4.19LEE691 pKa = 4.59SVEE694 pKa = 4.41
MM1 pKa = 7.85SYY3 pKa = 10.93QLFDD7 pKa = 3.66QLTGPLANVLPNFKK21 pKa = 10.55YY22 pKa = 9.93LAKK25 pKa = 9.13TVKK28 pKa = 10.32YY29 pKa = 10.04GIKK32 pKa = 10.05ADD34 pKa = 3.41VDD36 pKa = 3.31IGVTSEE42 pKa = 3.62FTNAYY47 pKa = 8.5KK48 pKa = 10.55RR49 pKa = 11.84RR50 pKa = 11.84GALFTNVLSPFGVFNAEE67 pKa = 4.27VVRR70 pKa = 11.84DD71 pKa = 3.8PPIFDD76 pKa = 5.18GINARR81 pKa = 11.84FFNDD85 pKa = 2.91YY86 pKa = 11.56GEE88 pKa = 4.14IDD90 pKa = 3.41EE91 pKa = 4.6RR92 pKa = 11.84AIAEE96 pKa = 4.23TLRR99 pKa = 11.84LKK101 pKa = 9.56GTYY104 pKa = 9.06RR105 pKa = 11.84PNVIAEE111 pKa = 4.65LAPTIASCNSEE122 pKa = 3.75NHH124 pKa = 5.34VSVLVNLLRR133 pKa = 11.84FSIIKK138 pKa = 10.46RR139 pKa = 11.84SDD141 pKa = 3.1PKK143 pKa = 11.03DD144 pKa = 3.64FKK146 pKa = 11.07FVNDD150 pKa = 4.16DD151 pKa = 3.41LFYY154 pKa = 11.14DD155 pKa = 4.35DD156 pKa = 4.02GHH158 pKa = 7.31LAVSYY163 pKa = 11.0KK164 pKa = 10.84NYY166 pKa = 10.59LKK168 pKa = 10.82RR169 pKa = 11.84EE170 pKa = 4.09FNEE173 pKa = 3.52EE174 pKa = 3.6LVFIFRR180 pKa = 11.84EE181 pKa = 5.32GIQIEE186 pKa = 4.4RR187 pKa = 11.84FSEE190 pKa = 4.3STVVGDD196 pKa = 6.22DD197 pKa = 4.42FYY199 pKa = 11.75CPNIEE204 pKa = 4.02NLSARR209 pKa = 11.84EE210 pKa = 3.84FNILLTLLGSIKK222 pKa = 10.14CDD224 pKa = 3.37YY225 pKa = 10.06PLRR228 pKa = 11.84LAFSTPRR235 pKa = 11.84LVDD238 pKa = 3.54KK239 pKa = 11.01LVLPLTTKK247 pKa = 9.58HH248 pKa = 5.54ASYY251 pKa = 11.2YY252 pKa = 10.38EE253 pKa = 4.05VVNDD257 pKa = 3.82FTANEE262 pKa = 3.73VDD264 pKa = 3.68QIIRR268 pKa = 11.84KK269 pKa = 9.48YY270 pKa = 9.76IFANRR275 pKa = 11.84VEE277 pKa = 4.39TAFDD281 pKa = 3.65LAYY284 pKa = 10.49LIVVTAMFAPLPRR297 pKa = 11.84AIEE300 pKa = 4.1ANGWVSPVNLIKK312 pKa = 10.77LPNVGSVRR320 pKa = 11.84GLMPEE325 pKa = 3.91MTGGQPLSKK334 pKa = 10.32SPQKK338 pKa = 11.1ALTWLNYY345 pKa = 8.51KK346 pKa = 10.07QSPGRR351 pKa = 11.84IAIHH355 pKa = 7.13AIAACEE361 pKa = 3.6AYY363 pKa = 8.82YY364 pKa = 10.45TGFFEE369 pKa = 5.9VLTASPNGIEE379 pKa = 4.42DD380 pKa = 3.98TLTAIGVSSSVTATPYY396 pKa = 11.59KK397 pKa = 9.74MFCEE401 pKa = 4.08SAAYY405 pKa = 10.19RR406 pKa = 11.84FGKK409 pKa = 10.35EE410 pKa = 4.46FDD412 pKa = 5.32LIWLTNAGVDD422 pKa = 4.14CYY424 pKa = 11.54SHH426 pKa = 7.26LLATEE431 pKa = 4.39PAALSILAKK440 pKa = 10.72VVDD443 pKa = 4.39TEE445 pKa = 4.4LEE447 pKa = 4.4GYY449 pKa = 9.94DD450 pKa = 3.96VYY452 pKa = 9.28TTSAFGEE459 pKa = 4.46DD460 pKa = 3.59TVHH463 pKa = 7.31VITKK467 pKa = 7.57EE468 pKa = 3.95VKK470 pKa = 8.6PALFPVLSMGINDD483 pKa = 4.24DD484 pKa = 4.18RR485 pKa = 11.84YY486 pKa = 9.19FANSLEE492 pKa = 4.24YY493 pKa = 10.7SATLYY498 pKa = 10.83ADD500 pKa = 3.19HH501 pKa = 7.12SEE503 pKa = 4.28GVLATTEE510 pKa = 3.99SDD512 pKa = 3.68HH513 pKa = 6.18MNKK516 pKa = 9.98ALSILRR522 pKa = 11.84IGGYY526 pKa = 9.84DD527 pKa = 3.0AVMTDD532 pKa = 3.38RR533 pKa = 11.84LSGLSYY539 pKa = 10.92RR540 pKa = 11.84NWAANSNGQVLPMLPPKK557 pKa = 10.67LKK559 pKa = 10.67GKK561 pKa = 7.74STYY564 pKa = 10.36VIPFNTIVKK573 pKa = 10.06RR574 pKa = 11.84KK575 pKa = 8.63KK576 pKa = 9.55SWMEE580 pKa = 3.76LGSLTDD586 pKa = 3.83SVTMTAKK593 pKa = 10.26INLRR597 pKa = 11.84GYY599 pKa = 11.24VIMTNGKK606 pKa = 9.09LVSTYY611 pKa = 10.29LPKK614 pKa = 10.47YY615 pKa = 7.52QTINVYY621 pKa = 10.03PKK623 pKa = 10.42SLNKK627 pKa = 10.36EE628 pKa = 3.7EE629 pKa = 4.11MVAVEE634 pKa = 4.74MKK636 pKa = 10.44TSATTIKK643 pKa = 10.23YY644 pKa = 9.64RR645 pKa = 11.84YY646 pKa = 9.88SGFLLAASQVEE657 pKa = 4.3APPARR662 pKa = 11.84LQLLSTDD669 pKa = 3.59ILGSQPIEE677 pKa = 4.16TSSEE681 pKa = 3.7EE682 pKa = 3.93AGAVGDD688 pKa = 4.16DD689 pKa = 4.19LEE691 pKa = 4.59SVEE694 pKa = 4.41
Molecular weight: 77.11 kDa
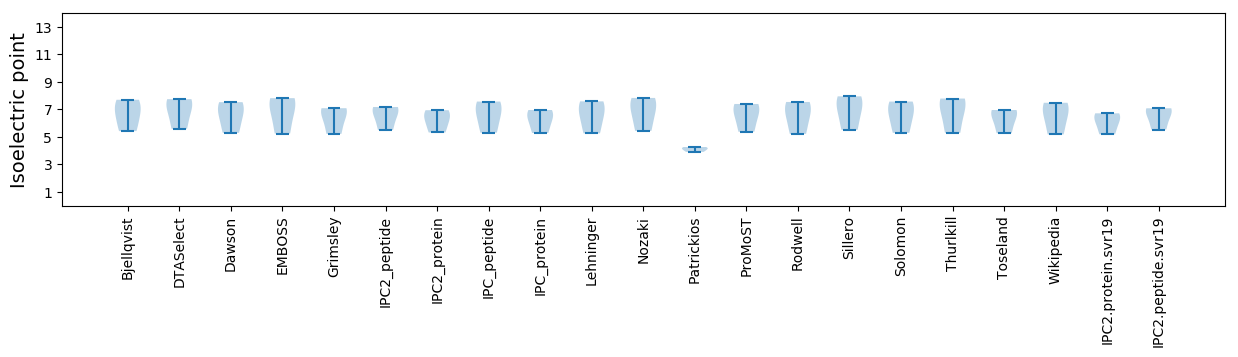
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4MEM7|M4MEM7_9VIRU Isoform of M4MG99 RNA-directed RNA polymerase (Fragment) OS=Scheffersomyces segobiensis virus L OX=1300323 GN=pol PE=3 SV=1
II1 pKa = 7.25PVFGFFVGCEE11 pKa = 4.08SSGSTTCPFTIIVYY25 pKa = 6.79GHH27 pKa = 6.84PWVAAHH33 pKa = 6.91RR34 pKa = 11.84NIEE37 pKa = 3.98RR38 pKa = 11.84RR39 pKa = 11.84GWCSGRR45 pKa = 11.84RR46 pKa = 11.84LGEE49 pKa = 3.86CRR51 pKa = 11.84MSGLNYY57 pKa = 9.86HH58 pKa = 6.43NPVPLYY64 pKa = 10.4VDD66 pKa = 3.53EE67 pKa = 5.03NMAGTVPEE75 pKa = 3.94KK76 pKa = 10.46SRR78 pKa = 11.84YY79 pKa = 8.85VLVHH83 pKa = 5.76VSNDD87 pKa = 3.3DD88 pKa = 3.91VPVHH92 pKa = 6.36NIDD95 pKa = 3.53YY96 pKa = 10.72PMLIPYY102 pKa = 9.62CGEE105 pKa = 3.63YY106 pKa = 10.39TIGKK110 pKa = 9.01RR111 pKa = 11.84IKK113 pKa = 10.16RR114 pKa = 11.84RR115 pKa = 11.84NIVCWYY121 pKa = 10.0AKK123 pKa = 10.51LDD125 pKa = 4.25FISIYY130 pKa = 8.29TPKK133 pKa = 10.57LMSIVGGYY141 pKa = 9.64ISGNIRR147 pKa = 11.84SGMTVEE153 pKa = 4.02NHH155 pKa = 6.3RR156 pKa = 11.84DD157 pKa = 3.23FYY159 pKa = 11.27MEE161 pKa = 3.84FDD163 pKa = 3.22KK164 pKa = 11.69VIFNKK169 pKa = 8.5VTSDD173 pKa = 3.88EE174 pKa = 3.91IRR176 pKa = 11.84EE177 pKa = 4.02LNEE180 pKa = 3.65YY181 pKa = 9.64VRR183 pKa = 11.84SLDD186 pKa = 3.39VTKK189 pKa = 9.86ITGDD193 pKa = 2.74HH194 pKa = 6.36HH195 pKa = 7.13LFVNRR200 pKa = 11.84QEE202 pKa = 4.07VDD204 pKa = 3.36TLLEE208 pKa = 3.79EE209 pKa = 4.44AMNLGKK215 pKa = 11.01YY216 pKa = 9.07NDD218 pKa = 3.77RR219 pKa = 11.84LAALMVLKK227 pKa = 10.66DD228 pKa = 3.34VASRR232 pKa = 11.84IGIDD236 pKa = 3.31NRR238 pKa = 11.84VSFITHH244 pKa = 6.54LVYY247 pKa = 10.6ILIAPPHH254 pKa = 6.24AFVHH258 pKa = 6.04LLSLIGEE265 pKa = 4.43STDD268 pKa = 3.46AEE270 pKa = 4.59DD271 pKa = 3.59YY272 pKa = 11.46ASILKK277 pKa = 10.27RR278 pKa = 11.84EE279 pKa = 4.08SGAAKK284 pKa = 10.0QLQTVFRR291 pKa = 11.84SDD293 pKa = 3.26LASLYY298 pKa = 10.04EE299 pKa = 4.17LNVLVNRR306 pKa = 11.84VDD308 pKa = 5.51SVVDD312 pKa = 3.34WNKK315 pKa = 10.87EE316 pKa = 3.14IDD318 pKa = 3.12HH319 pKa = 6.17RR320 pKa = 11.84TNVKK324 pKa = 10.16AVGISTEE331 pKa = 4.09DD332 pKa = 3.39VYY334 pKa = 11.4KK335 pKa = 10.69ISTEE339 pKa = 3.69IFKK342 pKa = 10.92DD343 pKa = 3.52AVRR346 pKa = 11.84EE347 pKa = 3.92GRR349 pKa = 11.84YY350 pKa = 7.69PQRR353 pKa = 11.84LEE355 pKa = 3.83WQDD358 pKa = 3.05YY359 pKa = 7.84WVQRR363 pKa = 11.84WSAMPNGSVISQYY376 pKa = 11.52DD377 pKa = 3.28EE378 pKa = 5.6DD379 pKa = 3.94IAIKK383 pKa = 10.52KK384 pKa = 8.79LLPFSARR391 pKa = 11.84VKK393 pKa = 10.2SAWFAAKK400 pKa = 9.63TEE402 pKa = 4.34SNYY405 pKa = 11.01DD406 pKa = 3.27YY407 pKa = 10.34WLKK410 pKa = 9.87RR411 pKa = 11.84TPQIYY416 pKa = 10.24ASTSTKK422 pKa = 10.45YY423 pKa = 9.85EE424 pKa = 3.76WGKK427 pKa = 10.21VRR429 pKa = 11.84ALYY432 pKa = 10.68GCDD435 pKa = 2.86VTSFLHH441 pKa = 6.88SDD443 pKa = 3.8FGMTNCEE450 pKa = 4.99DD451 pKa = 3.48ILPSYY456 pKa = 10.42FPVGRR461 pKa = 11.84KK462 pKa = 9.44ANEE465 pKa = 3.86KK466 pKa = 9.51YY467 pKa = 10.31VRR469 pKa = 11.84EE470 pKa = 4.27SIEE473 pKa = 4.04KK474 pKa = 9.79FRR476 pKa = 11.84EE477 pKa = 4.08TVPVCFDD484 pKa = 3.45YY485 pKa = 11.46DD486 pKa = 3.86DD487 pKa = 5.49FNSQHH492 pKa = 5.86STSSMRR498 pKa = 11.84TVLQAWLDD506 pKa = 3.81VNQKK510 pKa = 10.18YY511 pKa = 7.77LTEE514 pKa = 4.04EE515 pKa = 3.91QVNSVEE521 pKa = 4.13WTRR524 pKa = 11.84DD525 pKa = 3.41SLDD528 pKa = 3.88SILARR533 pKa = 11.84FNSSGDD539 pKa = 3.8TISLSGTLMSGWRR552 pKa = 11.84LTSFMNTVLNRR563 pKa = 11.84VYY565 pKa = 10.48LVKK568 pKa = 11.04ANLFKK573 pKa = 10.91HH574 pKa = 6.23LVYY577 pKa = 10.71ALHH580 pKa = 6.7NGDD583 pKa = 4.85DD584 pKa = 3.93MFGGAVNMAKK594 pKa = 10.15AVAIIRR600 pKa = 11.84DD601 pKa = 3.53AKK603 pKa = 10.78AIGVRR608 pKa = 11.84AQVSKK613 pKa = 10.17TNLGTIGEE621 pKa = 4.4FLRR624 pKa = 11.84VDD626 pKa = 3.56TRR628 pKa = 11.84AIDD631 pKa = 3.66PTNAQYY637 pKa = 11.36LSRR640 pKa = 11.84SVATLVHH647 pKa = 5.98GRR649 pKa = 11.84VEE651 pKa = 4.35SSAPNDD657 pKa = 3.29LRR659 pKa = 11.84EE660 pKa = 4.24LVSAIITRR668 pKa = 11.84SEE670 pKa = 3.91EE671 pKa = 4.1VISRR675 pKa = 11.84GGDD678 pKa = 3.03FRR680 pKa = 11.84LINKK684 pKa = 7.75LTQKK688 pKa = 9.4TLDD691 pKa = 3.26FAARR695 pKa = 11.84LFNVDD700 pKa = 2.99NTIIDD705 pKa = 3.92TMLSNHH711 pKa = 6.32PVQGGMNKK719 pKa = 9.88DD720 pKa = 3.46APVHH724 pKa = 5.48LWRR727 pKa = 11.84IEE729 pKa = 3.67RR730 pKa = 11.84RR731 pKa = 11.84TEE733 pKa = 3.54PFEE736 pKa = 5.75RR737 pKa = 11.84EE738 pKa = 3.8TLLARR743 pKa = 11.84YY744 pKa = 9.62SLIAPGLFNYY754 pKa = 9.67VDD756 pKa = 3.45RR757 pKa = 11.84VKK759 pKa = 10.86EE760 pKa = 3.8QFGVSEE766 pKa = 3.97NLIDD770 pKa = 5.33KK771 pKa = 11.01EE772 pKa = 4.09EE773 pKa = 4.0LLYY776 pKa = 10.78RR777 pKa = 11.84AGVSLEE783 pKa = 3.71KK784 pKa = 10.84SRR786 pKa = 11.84VYY788 pKa = 10.82YY789 pKa = 10.23EE790 pKa = 4.35LVIEE794 pKa = 4.57TTLGMDD800 pKa = 4.31IYY802 pKa = 10.68RR803 pKa = 11.84ALHH806 pKa = 5.78GVWKK810 pKa = 10.42SPGYY814 pKa = 9.15VAPIAKK820 pKa = 9.37IRR822 pKa = 11.84SLGLIPAKK830 pKa = 9.05EE831 pKa = 3.9LRR833 pKa = 11.84NLNSVPAHH841 pKa = 7.24LIRR844 pKa = 11.84TARR847 pKa = 11.84NPILMMGTIFF857 pKa = 4.87
II1 pKa = 7.25PVFGFFVGCEE11 pKa = 4.08SSGSTTCPFTIIVYY25 pKa = 6.79GHH27 pKa = 6.84PWVAAHH33 pKa = 6.91RR34 pKa = 11.84NIEE37 pKa = 3.98RR38 pKa = 11.84RR39 pKa = 11.84GWCSGRR45 pKa = 11.84RR46 pKa = 11.84LGEE49 pKa = 3.86CRR51 pKa = 11.84MSGLNYY57 pKa = 9.86HH58 pKa = 6.43NPVPLYY64 pKa = 10.4VDD66 pKa = 3.53EE67 pKa = 5.03NMAGTVPEE75 pKa = 3.94KK76 pKa = 10.46SRR78 pKa = 11.84YY79 pKa = 8.85VLVHH83 pKa = 5.76VSNDD87 pKa = 3.3DD88 pKa = 3.91VPVHH92 pKa = 6.36NIDD95 pKa = 3.53YY96 pKa = 10.72PMLIPYY102 pKa = 9.62CGEE105 pKa = 3.63YY106 pKa = 10.39TIGKK110 pKa = 9.01RR111 pKa = 11.84IKK113 pKa = 10.16RR114 pKa = 11.84RR115 pKa = 11.84NIVCWYY121 pKa = 10.0AKK123 pKa = 10.51LDD125 pKa = 4.25FISIYY130 pKa = 8.29TPKK133 pKa = 10.57LMSIVGGYY141 pKa = 9.64ISGNIRR147 pKa = 11.84SGMTVEE153 pKa = 4.02NHH155 pKa = 6.3RR156 pKa = 11.84DD157 pKa = 3.23FYY159 pKa = 11.27MEE161 pKa = 3.84FDD163 pKa = 3.22KK164 pKa = 11.69VIFNKK169 pKa = 8.5VTSDD173 pKa = 3.88EE174 pKa = 3.91IRR176 pKa = 11.84EE177 pKa = 4.02LNEE180 pKa = 3.65YY181 pKa = 9.64VRR183 pKa = 11.84SLDD186 pKa = 3.39VTKK189 pKa = 9.86ITGDD193 pKa = 2.74HH194 pKa = 6.36HH195 pKa = 7.13LFVNRR200 pKa = 11.84QEE202 pKa = 4.07VDD204 pKa = 3.36TLLEE208 pKa = 3.79EE209 pKa = 4.44AMNLGKK215 pKa = 11.01YY216 pKa = 9.07NDD218 pKa = 3.77RR219 pKa = 11.84LAALMVLKK227 pKa = 10.66DD228 pKa = 3.34VASRR232 pKa = 11.84IGIDD236 pKa = 3.31NRR238 pKa = 11.84VSFITHH244 pKa = 6.54LVYY247 pKa = 10.6ILIAPPHH254 pKa = 6.24AFVHH258 pKa = 6.04LLSLIGEE265 pKa = 4.43STDD268 pKa = 3.46AEE270 pKa = 4.59DD271 pKa = 3.59YY272 pKa = 11.46ASILKK277 pKa = 10.27RR278 pKa = 11.84EE279 pKa = 4.08SGAAKK284 pKa = 10.0QLQTVFRR291 pKa = 11.84SDD293 pKa = 3.26LASLYY298 pKa = 10.04EE299 pKa = 4.17LNVLVNRR306 pKa = 11.84VDD308 pKa = 5.51SVVDD312 pKa = 3.34WNKK315 pKa = 10.87EE316 pKa = 3.14IDD318 pKa = 3.12HH319 pKa = 6.17RR320 pKa = 11.84TNVKK324 pKa = 10.16AVGISTEE331 pKa = 4.09DD332 pKa = 3.39VYY334 pKa = 11.4KK335 pKa = 10.69ISTEE339 pKa = 3.69IFKK342 pKa = 10.92DD343 pKa = 3.52AVRR346 pKa = 11.84EE347 pKa = 3.92GRR349 pKa = 11.84YY350 pKa = 7.69PQRR353 pKa = 11.84LEE355 pKa = 3.83WQDD358 pKa = 3.05YY359 pKa = 7.84WVQRR363 pKa = 11.84WSAMPNGSVISQYY376 pKa = 11.52DD377 pKa = 3.28EE378 pKa = 5.6DD379 pKa = 3.94IAIKK383 pKa = 10.52KK384 pKa = 8.79LLPFSARR391 pKa = 11.84VKK393 pKa = 10.2SAWFAAKK400 pKa = 9.63TEE402 pKa = 4.34SNYY405 pKa = 11.01DD406 pKa = 3.27YY407 pKa = 10.34WLKK410 pKa = 9.87RR411 pKa = 11.84TPQIYY416 pKa = 10.24ASTSTKK422 pKa = 10.45YY423 pKa = 9.85EE424 pKa = 3.76WGKK427 pKa = 10.21VRR429 pKa = 11.84ALYY432 pKa = 10.68GCDD435 pKa = 2.86VTSFLHH441 pKa = 6.88SDD443 pKa = 3.8FGMTNCEE450 pKa = 4.99DD451 pKa = 3.48ILPSYY456 pKa = 10.42FPVGRR461 pKa = 11.84KK462 pKa = 9.44ANEE465 pKa = 3.86KK466 pKa = 9.51YY467 pKa = 10.31VRR469 pKa = 11.84EE470 pKa = 4.27SIEE473 pKa = 4.04KK474 pKa = 9.79FRR476 pKa = 11.84EE477 pKa = 4.08TVPVCFDD484 pKa = 3.45YY485 pKa = 11.46DD486 pKa = 3.86DD487 pKa = 5.49FNSQHH492 pKa = 5.86STSSMRR498 pKa = 11.84TVLQAWLDD506 pKa = 3.81VNQKK510 pKa = 10.18YY511 pKa = 7.77LTEE514 pKa = 4.04EE515 pKa = 3.91QVNSVEE521 pKa = 4.13WTRR524 pKa = 11.84DD525 pKa = 3.41SLDD528 pKa = 3.88SILARR533 pKa = 11.84FNSSGDD539 pKa = 3.8TISLSGTLMSGWRR552 pKa = 11.84LTSFMNTVLNRR563 pKa = 11.84VYY565 pKa = 10.48LVKK568 pKa = 11.04ANLFKK573 pKa = 10.91HH574 pKa = 6.23LVYY577 pKa = 10.71ALHH580 pKa = 6.7NGDD583 pKa = 4.85DD584 pKa = 3.93MFGGAVNMAKK594 pKa = 10.15AVAIIRR600 pKa = 11.84DD601 pKa = 3.53AKK603 pKa = 10.78AIGVRR608 pKa = 11.84AQVSKK613 pKa = 10.17TNLGTIGEE621 pKa = 4.4FLRR624 pKa = 11.84VDD626 pKa = 3.56TRR628 pKa = 11.84AIDD631 pKa = 3.66PTNAQYY637 pKa = 11.36LSRR640 pKa = 11.84SVATLVHH647 pKa = 5.98GRR649 pKa = 11.84VEE651 pKa = 4.35SSAPNDD657 pKa = 3.29LRR659 pKa = 11.84EE660 pKa = 4.24LVSAIITRR668 pKa = 11.84SEE670 pKa = 3.91EE671 pKa = 4.1VISRR675 pKa = 11.84GGDD678 pKa = 3.03FRR680 pKa = 11.84LINKK684 pKa = 7.75LTQKK688 pKa = 9.4TLDD691 pKa = 3.26FAARR695 pKa = 11.84LFNVDD700 pKa = 2.99NTIIDD705 pKa = 3.92TMLSNHH711 pKa = 6.32PVQGGMNKK719 pKa = 9.88DD720 pKa = 3.46APVHH724 pKa = 5.48LWRR727 pKa = 11.84IEE729 pKa = 3.67RR730 pKa = 11.84RR731 pKa = 11.84TEE733 pKa = 3.54PFEE736 pKa = 5.75RR737 pKa = 11.84EE738 pKa = 3.8TLLARR743 pKa = 11.84YY744 pKa = 9.62SLIAPGLFNYY754 pKa = 9.67VDD756 pKa = 3.45RR757 pKa = 11.84VKK759 pKa = 10.86EE760 pKa = 3.8QFGVSEE766 pKa = 3.97NLIDD770 pKa = 5.33KK771 pKa = 11.01EE772 pKa = 4.09EE773 pKa = 4.0LLYY776 pKa = 10.78RR777 pKa = 11.84AGVSLEE783 pKa = 3.71KK784 pKa = 10.84SRR786 pKa = 11.84VYY788 pKa = 10.82YY789 pKa = 10.23EE790 pKa = 4.35LVIEE794 pKa = 4.57TTLGMDD800 pKa = 4.31IYY802 pKa = 10.68RR803 pKa = 11.84ALHH806 pKa = 5.78GVWKK810 pKa = 10.42SPGYY814 pKa = 9.15VAPIAKK820 pKa = 9.37IRR822 pKa = 11.84SLGLIPAKK830 pKa = 9.05EE831 pKa = 3.9LRR833 pKa = 11.84NLNSVPAHH841 pKa = 7.24LIRR844 pKa = 11.84TARR847 pKa = 11.84NPILMMGTIFF857 pKa = 4.87
Molecular weight: 97.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3051 |
694 |
1500 |
1017.0 |
114.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.211 ± 0.445 | 0.983 ± 0.039 |
5.539 ± 0.13 | 5.965 ± 0.076 |
4.326 ± 0.211 | 5.637 ± 0.029 |
1.901 ± 0.262 | 6.391 ± 0.173 |
5.179 ± 0.148 | 9.407 ± 0.278 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.229 ± 0.068 | 5.375 ± 0.056 |
4.064 ± 0.284 | 1.803 ± 0.065 |
5.867 ± 0.593 | 7.407 ± 0.064 |
6.227 ± 0.265 | 8.325 ± 0.195 |
1.311 ± 0.208 | 4.851 ± 0.11 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
