
Human adenovirus sp.
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus; unclassified Human adenoviruses
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
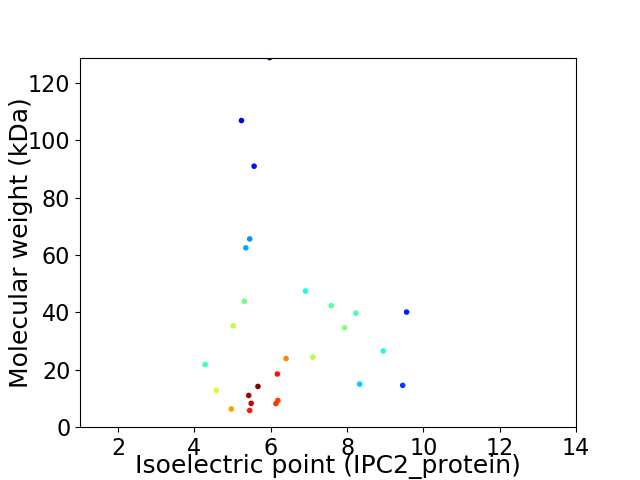
Virtual 2D-PAGE plot for 27 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6HVP9|A0A1W6HVP9_9ADEN E3-19K OS=Human adenovirus sp. OX=1907210 PE=3 SV=1
MM1 pKa = 7.34RR2 pKa = 11.84HH3 pKa = 6.25LRR5 pKa = 11.84FLPQEE10 pKa = 4.33IISAEE15 pKa = 4.03TGNEE19 pKa = 3.27ILEE22 pKa = 4.52FVVHH26 pKa = 6.97ALMGDD31 pKa = 3.71DD32 pKa = 4.84PEE34 pKa = 5.93PPVQLFEE41 pKa = 4.7PPTLQEE47 pKa = 4.5LYY49 pKa = 10.31DD50 pKa = 4.17LEE52 pKa = 4.77VEE54 pKa = 4.35GSEE57 pKa = 4.86DD58 pKa = 3.8SNEE61 pKa = 3.83EE62 pKa = 3.77AVNGFFTDD70 pKa = 3.97SMLLAANEE78 pKa = 4.13GLEE81 pKa = 4.61LDD83 pKa = 4.47PPLDD87 pKa = 3.81TFDD90 pKa = 3.65TPGVIVEE97 pKa = 4.52SGTGVRR103 pKa = 11.84KK104 pKa = 9.9LPDD107 pKa = 4.4LGSVDD112 pKa = 4.89CDD114 pKa = 3.2LHH116 pKa = 8.32CYY118 pKa = 10.19EE119 pKa = 5.87DD120 pKa = 4.7GFPLSDD126 pKa = 3.31EE127 pKa = 4.09EE128 pKa = 5.17DD129 pKa = 3.58RR130 pKa = 11.84EE131 pKa = 4.4KK132 pKa = 11.14EE133 pKa = 4.08QFMQTAAGEE142 pKa = 4.52GVKK145 pKa = 10.16AASVGFQLDD154 pKa = 3.96CPEE157 pKa = 4.73LPGHH161 pKa = 5.76GCKK164 pKa = 10.06SCEE167 pKa = 3.93FHH169 pKa = 7.55RR170 pKa = 11.84KK171 pKa = 6.24NTGVKK176 pKa = 9.89EE177 pKa = 4.01LLCSLCYY184 pKa = 9.86MRR186 pKa = 11.84AHH188 pKa = 6.47CHH190 pKa = 6.56FIYY193 pKa = 10.49SKK195 pKa = 10.92CVV197 pKa = 2.88
MM1 pKa = 7.34RR2 pKa = 11.84HH3 pKa = 6.25LRR5 pKa = 11.84FLPQEE10 pKa = 4.33IISAEE15 pKa = 4.03TGNEE19 pKa = 3.27ILEE22 pKa = 4.52FVVHH26 pKa = 6.97ALMGDD31 pKa = 3.71DD32 pKa = 4.84PEE34 pKa = 5.93PPVQLFEE41 pKa = 4.7PPTLQEE47 pKa = 4.5LYY49 pKa = 10.31DD50 pKa = 4.17LEE52 pKa = 4.77VEE54 pKa = 4.35GSEE57 pKa = 4.86DD58 pKa = 3.8SNEE61 pKa = 3.83EE62 pKa = 3.77AVNGFFTDD70 pKa = 3.97SMLLAANEE78 pKa = 4.13GLEE81 pKa = 4.61LDD83 pKa = 4.47PPLDD87 pKa = 3.81TFDD90 pKa = 3.65TPGVIVEE97 pKa = 4.52SGTGVRR103 pKa = 11.84KK104 pKa = 9.9LPDD107 pKa = 4.4LGSVDD112 pKa = 4.89CDD114 pKa = 3.2LHH116 pKa = 8.32CYY118 pKa = 10.19EE119 pKa = 5.87DD120 pKa = 4.7GFPLSDD126 pKa = 3.31EE127 pKa = 4.09EE128 pKa = 5.17DD129 pKa = 3.58RR130 pKa = 11.84EE131 pKa = 4.4KK132 pKa = 11.14EE133 pKa = 4.08QFMQTAAGEE142 pKa = 4.52GVKK145 pKa = 10.16AASVGFQLDD154 pKa = 3.96CPEE157 pKa = 4.73LPGHH161 pKa = 5.76GCKK164 pKa = 10.06SCEE167 pKa = 3.93FHH169 pKa = 7.55RR170 pKa = 11.84KK171 pKa = 6.24NTGVKK176 pKa = 9.89EE177 pKa = 4.01LLCSLCYY184 pKa = 9.86MRR186 pKa = 11.84AHH188 pKa = 6.47CHH190 pKa = 6.56FIYY193 pKa = 10.49SKK195 pKa = 10.92CVV197 pKa = 2.88
Molecular weight: 21.83 kDa
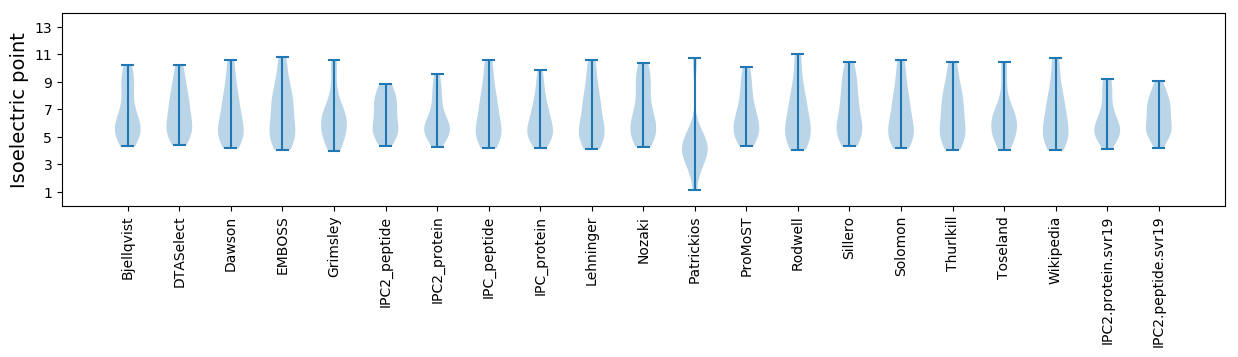
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6HVP8|A0A1W6HVP8_9ADEN Early E1A protein OS=Human adenovirus sp. OX=1907210 PE=3 SV=1
MM1 pKa = 7.7SKK3 pKa = 10.43RR4 pKa = 11.84KK5 pKa = 9.85YY6 pKa = 9.79KK7 pKa = 10.9EE8 pKa = 3.54EE9 pKa = 3.89MLQVIAPEE17 pKa = 4.73VYY19 pKa = 9.76GQPLKK24 pKa = 10.9DD25 pKa = 3.29EE26 pKa = 4.48KK27 pKa = 10.91KK28 pKa = 8.53PRR30 pKa = 11.84KK31 pKa = 9.09IKK33 pKa = 10.18RR34 pKa = 11.84AKK36 pKa = 9.47KK37 pKa = 9.74DD38 pKa = 3.34KK39 pKa = 10.97KK40 pKa = 10.65EE41 pKa = 4.15EE42 pKa = 3.99EE43 pKa = 4.72DD44 pKa = 4.04GDD46 pKa = 4.23DD47 pKa = 3.73GLAEE51 pKa = 4.4FVRR54 pKa = 11.84EE55 pKa = 3.86FAPRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84VQWRR65 pKa = 11.84GRR67 pKa = 11.84KK68 pKa = 8.6VRR70 pKa = 11.84HH71 pKa = 5.92LLRR74 pKa = 11.84PGTSVVFTPGEE85 pKa = 3.87RR86 pKa = 11.84SSATFKK92 pKa = 10.65RR93 pKa = 11.84SYY95 pKa = 11.27DD96 pKa = 3.53EE97 pKa = 5.21VYY99 pKa = 11.03GDD101 pKa = 5.76DD102 pKa = 6.37DD103 pKa = 4.14ILEE106 pKa = 4.15QAADD110 pKa = 3.8RR111 pKa = 11.84LGEE114 pKa = 3.76FAYY117 pKa = 10.26GKK119 pKa = 10.08RR120 pKa = 11.84SRR122 pKa = 11.84INPKK126 pKa = 10.42DD127 pKa = 3.46EE128 pKa = 4.27TVSIPLDD135 pKa = 3.78HH136 pKa = 7.08GNPTPSLKK144 pKa = 10.31PVTLQQVLPVTPRR157 pKa = 11.84TGVKK161 pKa = 10.0RR162 pKa = 11.84EE163 pKa = 4.06GEE165 pKa = 3.96DD166 pKa = 4.36LYY168 pKa = 10.28PTMQLMVPKK177 pKa = 9.82RR178 pKa = 11.84QKK180 pKa = 11.1LEE182 pKa = 3.99DD183 pKa = 3.51VLEE186 pKa = 4.25KK187 pKa = 11.2VKK189 pKa = 10.61VDD191 pKa = 3.69PDD193 pKa = 3.28IQPEE197 pKa = 4.48VKK199 pKa = 9.97VRR201 pKa = 11.84PIKK204 pKa = 10.32QVAPGLGVQTVDD216 pKa = 2.93IKK218 pKa = 11.14IPTEE222 pKa = 3.9SMEE225 pKa = 4.38VQTEE229 pKa = 4.07PAKK232 pKa = 9.51PTATSTEE239 pKa = 4.22VQTDD243 pKa = 2.89PWMPMPITTDD253 pKa = 2.91AAGPTRR259 pKa = 11.84RR260 pKa = 11.84SRR262 pKa = 11.84RR263 pKa = 11.84KK264 pKa = 9.41YY265 pKa = 10.39GPASLLMPNYY275 pKa = 9.18VVHH278 pKa = 6.52PSIIPTPGYY287 pKa = 9.95RR288 pKa = 11.84GTRR291 pKa = 11.84YY292 pKa = 9.37YY293 pKa = 10.8RR294 pKa = 11.84SRR296 pKa = 11.84NSTSRR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84RR304 pKa = 11.84KK305 pKa = 7.96TPANRR310 pKa = 11.84SRR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84TSKK319 pKa = 8.65PTPGALVRR327 pKa = 11.84QVYY330 pKa = 9.83RR331 pKa = 11.84NSSAEE336 pKa = 4.0PLTLPRR342 pKa = 11.84ARR344 pKa = 11.84YY345 pKa = 8.46HH346 pKa = 6.34PSIITT351 pKa = 3.35
MM1 pKa = 7.7SKK3 pKa = 10.43RR4 pKa = 11.84KK5 pKa = 9.85YY6 pKa = 9.79KK7 pKa = 10.9EE8 pKa = 3.54EE9 pKa = 3.89MLQVIAPEE17 pKa = 4.73VYY19 pKa = 9.76GQPLKK24 pKa = 10.9DD25 pKa = 3.29EE26 pKa = 4.48KK27 pKa = 10.91KK28 pKa = 8.53PRR30 pKa = 11.84KK31 pKa = 9.09IKK33 pKa = 10.18RR34 pKa = 11.84AKK36 pKa = 9.47KK37 pKa = 9.74DD38 pKa = 3.34KK39 pKa = 10.97KK40 pKa = 10.65EE41 pKa = 4.15EE42 pKa = 3.99EE43 pKa = 4.72DD44 pKa = 4.04GDD46 pKa = 4.23DD47 pKa = 3.73GLAEE51 pKa = 4.4FVRR54 pKa = 11.84EE55 pKa = 3.86FAPRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84VQWRR65 pKa = 11.84GRR67 pKa = 11.84KK68 pKa = 8.6VRR70 pKa = 11.84HH71 pKa = 5.92LLRR74 pKa = 11.84PGTSVVFTPGEE85 pKa = 3.87RR86 pKa = 11.84SSATFKK92 pKa = 10.65RR93 pKa = 11.84SYY95 pKa = 11.27DD96 pKa = 3.53EE97 pKa = 5.21VYY99 pKa = 11.03GDD101 pKa = 5.76DD102 pKa = 6.37DD103 pKa = 4.14ILEE106 pKa = 4.15QAADD110 pKa = 3.8RR111 pKa = 11.84LGEE114 pKa = 3.76FAYY117 pKa = 10.26GKK119 pKa = 10.08RR120 pKa = 11.84SRR122 pKa = 11.84INPKK126 pKa = 10.42DD127 pKa = 3.46EE128 pKa = 4.27TVSIPLDD135 pKa = 3.78HH136 pKa = 7.08GNPTPSLKK144 pKa = 10.31PVTLQQVLPVTPRR157 pKa = 11.84TGVKK161 pKa = 10.0RR162 pKa = 11.84EE163 pKa = 4.06GEE165 pKa = 3.96DD166 pKa = 4.36LYY168 pKa = 10.28PTMQLMVPKK177 pKa = 9.82RR178 pKa = 11.84QKK180 pKa = 11.1LEE182 pKa = 3.99DD183 pKa = 3.51VLEE186 pKa = 4.25KK187 pKa = 11.2VKK189 pKa = 10.61VDD191 pKa = 3.69PDD193 pKa = 3.28IQPEE197 pKa = 4.48VKK199 pKa = 9.97VRR201 pKa = 11.84PIKK204 pKa = 10.32QVAPGLGVQTVDD216 pKa = 2.93IKK218 pKa = 11.14IPTEE222 pKa = 3.9SMEE225 pKa = 4.38VQTEE229 pKa = 4.07PAKK232 pKa = 9.51PTATSTEE239 pKa = 4.22VQTDD243 pKa = 2.89PWMPMPITTDD253 pKa = 2.91AAGPTRR259 pKa = 11.84RR260 pKa = 11.84SRR262 pKa = 11.84RR263 pKa = 11.84KK264 pKa = 9.41YY265 pKa = 10.39GPASLLMPNYY275 pKa = 9.18VVHH278 pKa = 6.52PSIIPTPGYY287 pKa = 9.95RR288 pKa = 11.84GTRR291 pKa = 11.84YY292 pKa = 9.37YY293 pKa = 10.8RR294 pKa = 11.84SRR296 pKa = 11.84NSTSRR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84RR304 pKa = 11.84KK305 pKa = 7.96TPANRR310 pKa = 11.84SRR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84TSKK319 pKa = 8.65PTPGALVRR327 pKa = 11.84QVYY330 pKa = 9.83RR331 pKa = 11.84NSSAEE336 pKa = 4.0PLTLPRR342 pKa = 11.84ARR344 pKa = 11.84YY345 pKa = 8.46HH346 pKa = 6.34PSIITT351 pKa = 3.35
Molecular weight: 40.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8516 |
54 |
1122 |
315.4 |
35.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.858 ± 0.329 | 1.973 ± 0.416 |
5.202 ± 0.197 | 6.247 ± 0.406 |
3.84 ± 0.243 | 6.118 ± 0.286 |
2.302 ± 0.238 | 4.357 ± 0.305 |
4.556 ± 0.393 | 9.382 ± 0.498 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.994 ± 0.255 | 4.979 ± 0.451 |
6.329 ± 0.36 | 4.439 ± 0.287 |
6.353 ± 0.413 | 6.881 ± 0.33 |
6.165 ± 0.457 | 6.118 ± 0.351 |
1.327 ± 0.131 | 3.581 ± 0.307 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
