
Liao ning virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Seadornavirus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
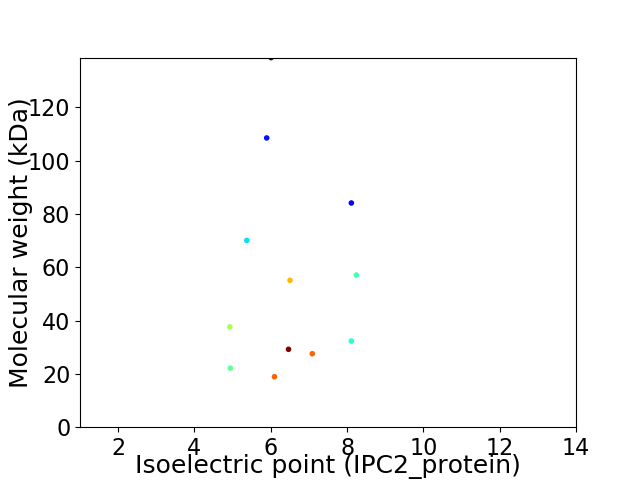
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2TV14|Q2TV14_9REOV VP6 OS=Liao ning virus OX=246280 PE=4 SV=1
MM1 pKa = 8.25DD2 pKa = 4.91IPSPNCGSVQKK13 pKa = 11.03NCFQVYY19 pKa = 9.93KK20 pKa = 9.59SQNVYY25 pKa = 9.86RR26 pKa = 11.84VRR28 pKa = 11.84FCVEE32 pKa = 3.85GKK34 pKa = 9.48IHH36 pKa = 7.16DD37 pKa = 4.05VPLPGVNTHH46 pKa = 6.49RR47 pKa = 11.84DD48 pKa = 3.05IIGTVKK54 pKa = 10.1YY55 pKa = 9.98FCEE58 pKa = 4.0LMGLNIVDD66 pKa = 3.48NSLRR70 pKa = 11.84LNWLNLSDD78 pKa = 4.07MKK80 pKa = 11.42NHH82 pKa = 6.66GDD84 pKa = 3.43NVTLAKK90 pKa = 10.5VGSEE94 pKa = 4.0YY95 pKa = 11.47GDD97 pKa = 3.92LFVKK101 pKa = 10.47KK102 pKa = 10.56LPTLPINIHH111 pKa = 4.28STNYY115 pKa = 9.6VFGKK119 pKa = 9.2YY120 pKa = 8.59SVFARR125 pKa = 11.84VHH127 pKa = 6.37GIVKK131 pKa = 10.36LLNDD135 pKa = 3.92DD136 pKa = 4.18NYY138 pKa = 11.49DD139 pKa = 3.38VGVILEE145 pKa = 4.25KK146 pKa = 10.95CNEE149 pKa = 3.81IRR151 pKa = 11.84VMTVNIIVMGLKK163 pKa = 10.34SLMDD167 pKa = 3.23SHH169 pKa = 7.48RR170 pKa = 11.84EE171 pKa = 3.99SGGTLHH177 pKa = 7.54GDD179 pKa = 3.36CTVQNLMCDD188 pKa = 3.08KK189 pKa = 10.92FGMLKK194 pKa = 10.55LVDD197 pKa = 4.57PGNLMSQTVIWKK209 pKa = 9.96NDD211 pKa = 2.98TQYY214 pKa = 11.37FGNTVDD220 pKa = 6.01DD221 pKa = 4.59EE222 pKa = 4.77VCSFVMACLKK232 pKa = 8.0TTANLRR238 pKa = 11.84NIEE241 pKa = 4.15TSEE244 pKa = 3.8LTIPFITDD252 pKa = 4.18LYY254 pKa = 11.44LLDD257 pKa = 4.48TVLSDD262 pKa = 3.75PNVIHH267 pKa = 6.44GTRR270 pKa = 11.84EE271 pKa = 4.22SGWLEE276 pKa = 3.94VGSDD280 pKa = 3.8FVSIVQMFVALANMVRR296 pKa = 11.84PYY298 pKa = 11.27NLDD301 pKa = 3.6DD302 pKa = 3.72VVSALKK308 pKa = 9.02TQVTGEE314 pKa = 4.26NEE316 pKa = 4.01TFSNVIEE323 pKa = 4.0KK324 pKa = 10.65HH325 pKa = 5.86EE326 pKa = 5.01DD327 pKa = 3.6GEE329 pKa = 4.81HH330 pKa = 6.87PDD332 pKa = 3.59SDD334 pKa = 4.72LEE336 pKa = 4.16
MM1 pKa = 8.25DD2 pKa = 4.91IPSPNCGSVQKK13 pKa = 11.03NCFQVYY19 pKa = 9.93KK20 pKa = 9.59SQNVYY25 pKa = 9.86RR26 pKa = 11.84VRR28 pKa = 11.84FCVEE32 pKa = 3.85GKK34 pKa = 9.48IHH36 pKa = 7.16DD37 pKa = 4.05VPLPGVNTHH46 pKa = 6.49RR47 pKa = 11.84DD48 pKa = 3.05IIGTVKK54 pKa = 10.1YY55 pKa = 9.98FCEE58 pKa = 4.0LMGLNIVDD66 pKa = 3.48NSLRR70 pKa = 11.84LNWLNLSDD78 pKa = 4.07MKK80 pKa = 11.42NHH82 pKa = 6.66GDD84 pKa = 3.43NVTLAKK90 pKa = 10.5VGSEE94 pKa = 4.0YY95 pKa = 11.47GDD97 pKa = 3.92LFVKK101 pKa = 10.47KK102 pKa = 10.56LPTLPINIHH111 pKa = 4.28STNYY115 pKa = 9.6VFGKK119 pKa = 9.2YY120 pKa = 8.59SVFARR125 pKa = 11.84VHH127 pKa = 6.37GIVKK131 pKa = 10.36LLNDD135 pKa = 3.92DD136 pKa = 4.18NYY138 pKa = 11.49DD139 pKa = 3.38VGVILEE145 pKa = 4.25KK146 pKa = 10.95CNEE149 pKa = 3.81IRR151 pKa = 11.84VMTVNIIVMGLKK163 pKa = 10.34SLMDD167 pKa = 3.23SHH169 pKa = 7.48RR170 pKa = 11.84EE171 pKa = 3.99SGGTLHH177 pKa = 7.54GDD179 pKa = 3.36CTVQNLMCDD188 pKa = 3.08KK189 pKa = 10.92FGMLKK194 pKa = 10.55LVDD197 pKa = 4.57PGNLMSQTVIWKK209 pKa = 9.96NDD211 pKa = 2.98TQYY214 pKa = 11.37FGNTVDD220 pKa = 6.01DD221 pKa = 4.59EE222 pKa = 4.77VCSFVMACLKK232 pKa = 8.0TTANLRR238 pKa = 11.84NIEE241 pKa = 4.15TSEE244 pKa = 3.8LTIPFITDD252 pKa = 4.18LYY254 pKa = 11.44LLDD257 pKa = 4.48TVLSDD262 pKa = 3.75PNVIHH267 pKa = 6.44GTRR270 pKa = 11.84EE271 pKa = 4.22SGWLEE276 pKa = 3.94VGSDD280 pKa = 3.8FVSIVQMFVALANMVRR296 pKa = 11.84PYY298 pKa = 11.27NLDD301 pKa = 3.6DD302 pKa = 3.72VVSALKK308 pKa = 9.02TQVTGEE314 pKa = 4.26NEE316 pKa = 4.01TFSNVIEE323 pKa = 4.0KK324 pKa = 10.65HH325 pKa = 5.86EE326 pKa = 5.01DD327 pKa = 3.6GEE329 pKa = 4.81HH330 pKa = 6.87PDD332 pKa = 3.59SDD334 pKa = 4.72LEE336 pKa = 4.16
Molecular weight: 37.64 kDa
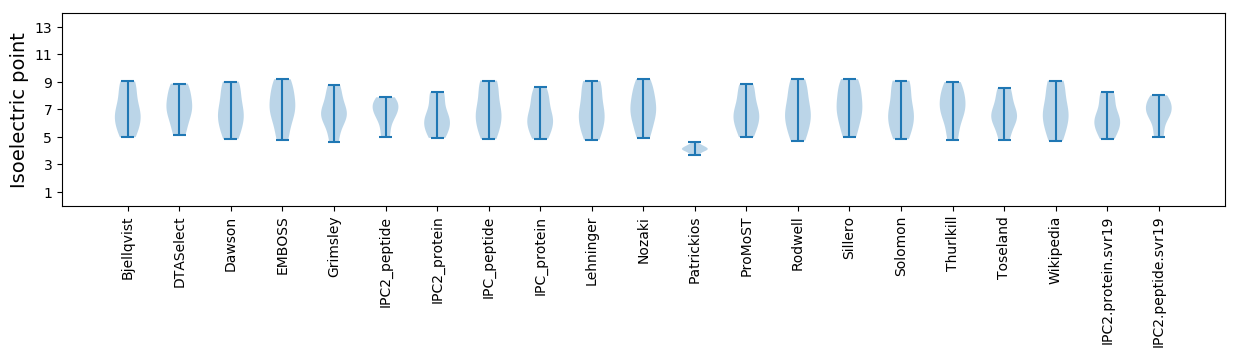
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2TV15|Q2TV15_9REOV VP5 OS=Liao ning virus OX=246280 PE=4 SV=1
MM1 pKa = 7.58SKK3 pKa = 10.79LSTIPRR9 pKa = 11.84TRR11 pKa = 11.84SQVVGLTSKK20 pKa = 11.01LKK22 pKa = 10.88DD23 pKa = 3.37FTKK26 pKa = 10.56SVEE29 pKa = 4.1KK30 pKa = 8.54TTLLTQSEE38 pKa = 4.68LKK40 pKa = 9.99KK41 pKa = 9.38QHH43 pKa = 5.96RR44 pKa = 11.84HH45 pKa = 5.67LLSQQPEE52 pKa = 4.58LFSGLAKK59 pKa = 10.68LYY61 pKa = 9.31TSNVSKK67 pKa = 10.63FEE69 pKa = 4.39RR70 pKa = 11.84IIRR73 pKa = 11.84LYY75 pKa = 10.38GGQGSKK81 pKa = 10.72HH82 pKa = 6.52SISSVGFSHH91 pKa = 8.01LIEE94 pKa = 4.25YY95 pKa = 9.52CNNYY99 pKa = 9.93DD100 pKa = 5.58LPDD103 pKa = 4.88DD104 pKa = 5.2DD105 pKa = 4.75STLKK109 pKa = 10.75AICFTIFQIHH119 pKa = 6.06SNEE122 pKa = 3.75LRR124 pKa = 11.84NGGANALFTIASEE137 pKa = 4.43YY138 pKa = 10.75FNAPAVKK145 pKa = 9.78EE146 pKa = 4.01IVGEE150 pKa = 3.98HH151 pKa = 5.25VKK153 pKa = 10.39IKK155 pKa = 10.36SCNKK159 pKa = 8.91VPVAIVNFKK168 pKa = 10.83ALTQIIPYY176 pKa = 9.67RR177 pKa = 11.84GVKK180 pKa = 9.79ALRR183 pKa = 11.84TKK185 pKa = 10.79LLEE188 pKa = 4.46TQDD191 pKa = 3.84GVALEE196 pKa = 5.09HH197 pKa = 6.87IGSFYY202 pKa = 9.92NTCKK206 pKa = 10.5INGEE210 pKa = 3.72FDD212 pKa = 3.45AVSFCGLKK220 pKa = 10.31LRR222 pKa = 11.84QSCIVSVAGHH232 pKa = 7.26AIDD235 pKa = 5.03VGPDD239 pKa = 3.08YY240 pKa = 10.54YY241 pKa = 11.0RR242 pKa = 11.84QMLAEE247 pKa = 4.18VTALLKK253 pKa = 10.64LKK255 pKa = 10.28KK256 pKa = 9.96KK257 pKa = 9.08VGSVVMHH264 pKa = 6.74GNNQFGFVKK273 pKa = 10.29PLDD276 pKa = 3.84ASQTQNVTQIRR287 pKa = 11.84KK288 pKa = 8.99ILNDD292 pKa = 3.32ASRR295 pKa = 11.84VSFINVRR302 pKa = 11.84NAASRR307 pKa = 11.84DD308 pKa = 3.65EE309 pKa = 4.21MVYY312 pKa = 10.46AYY314 pKa = 9.6DD315 pKa = 3.93PCLLYY320 pKa = 11.12YY321 pKa = 9.4ITIPNNGNKK330 pKa = 8.65SHH332 pKa = 7.52CIQALNLSGLLEE344 pKa = 4.05HH345 pKa = 7.19RR346 pKa = 11.84LLHH349 pKa = 6.25FGSLGTSCRR358 pKa = 11.84DD359 pKa = 3.15PSCRR363 pKa = 11.84VIGIDD368 pKa = 3.66RR369 pKa = 11.84PEE371 pKa = 4.75LYY373 pKa = 10.44SEE375 pKa = 4.69LNRR378 pKa = 11.84IVDD381 pKa = 3.37NTTIKK386 pKa = 10.82DD387 pKa = 3.33RR388 pKa = 11.84ADD390 pKa = 3.14AMLEE394 pKa = 3.87INADD398 pKa = 3.4EE399 pKa = 4.11MSSVKK404 pKa = 10.6SSGSGKK410 pKa = 10.43SLTKK414 pKa = 10.33GKK416 pKa = 9.48YY417 pKa = 8.57VRR419 pKa = 11.84NSIDD423 pKa = 3.06KK424 pKa = 10.4GHH426 pKa = 6.7NLTDD430 pKa = 3.63LGKK433 pKa = 10.71ANGDD437 pKa = 3.67QYY439 pKa = 11.77SHH441 pKa = 6.82YY442 pKa = 10.71SQMSKK447 pKa = 10.64PSGSEE452 pKa = 3.0ISKK455 pKa = 10.64FITAEE460 pKa = 3.88AVKK463 pKa = 10.44GISDD467 pKa = 3.78SSLDD471 pKa = 3.61ATEE474 pKa = 5.21LSLKK478 pKa = 10.57LAMLQSSAVTIGASKK493 pKa = 11.29NEE495 pKa = 4.14TKK497 pKa = 10.39PVEE500 pKa = 4.38TKK502 pKa = 10.04VVKK505 pKa = 10.55APSVNKK511 pKa = 9.05TADD514 pKa = 3.52EE515 pKa = 4.29LEE517 pKa = 4.58EE518 pKa = 4.05II519 pKa = 4.67
MM1 pKa = 7.58SKK3 pKa = 10.79LSTIPRR9 pKa = 11.84TRR11 pKa = 11.84SQVVGLTSKK20 pKa = 11.01LKK22 pKa = 10.88DD23 pKa = 3.37FTKK26 pKa = 10.56SVEE29 pKa = 4.1KK30 pKa = 8.54TTLLTQSEE38 pKa = 4.68LKK40 pKa = 9.99KK41 pKa = 9.38QHH43 pKa = 5.96RR44 pKa = 11.84HH45 pKa = 5.67LLSQQPEE52 pKa = 4.58LFSGLAKK59 pKa = 10.68LYY61 pKa = 9.31TSNVSKK67 pKa = 10.63FEE69 pKa = 4.39RR70 pKa = 11.84IIRR73 pKa = 11.84LYY75 pKa = 10.38GGQGSKK81 pKa = 10.72HH82 pKa = 6.52SISSVGFSHH91 pKa = 8.01LIEE94 pKa = 4.25YY95 pKa = 9.52CNNYY99 pKa = 9.93DD100 pKa = 5.58LPDD103 pKa = 4.88DD104 pKa = 5.2DD105 pKa = 4.75STLKK109 pKa = 10.75AICFTIFQIHH119 pKa = 6.06SNEE122 pKa = 3.75LRR124 pKa = 11.84NGGANALFTIASEE137 pKa = 4.43YY138 pKa = 10.75FNAPAVKK145 pKa = 9.78EE146 pKa = 4.01IVGEE150 pKa = 3.98HH151 pKa = 5.25VKK153 pKa = 10.39IKK155 pKa = 10.36SCNKK159 pKa = 8.91VPVAIVNFKK168 pKa = 10.83ALTQIIPYY176 pKa = 9.67RR177 pKa = 11.84GVKK180 pKa = 9.79ALRR183 pKa = 11.84TKK185 pKa = 10.79LLEE188 pKa = 4.46TQDD191 pKa = 3.84GVALEE196 pKa = 5.09HH197 pKa = 6.87IGSFYY202 pKa = 9.92NTCKK206 pKa = 10.5INGEE210 pKa = 3.72FDD212 pKa = 3.45AVSFCGLKK220 pKa = 10.31LRR222 pKa = 11.84QSCIVSVAGHH232 pKa = 7.26AIDD235 pKa = 5.03VGPDD239 pKa = 3.08YY240 pKa = 10.54YY241 pKa = 11.0RR242 pKa = 11.84QMLAEE247 pKa = 4.18VTALLKK253 pKa = 10.64LKK255 pKa = 10.28KK256 pKa = 9.96KK257 pKa = 9.08VGSVVMHH264 pKa = 6.74GNNQFGFVKK273 pKa = 10.29PLDD276 pKa = 3.84ASQTQNVTQIRR287 pKa = 11.84KK288 pKa = 8.99ILNDD292 pKa = 3.32ASRR295 pKa = 11.84VSFINVRR302 pKa = 11.84NAASRR307 pKa = 11.84DD308 pKa = 3.65EE309 pKa = 4.21MVYY312 pKa = 10.46AYY314 pKa = 9.6DD315 pKa = 3.93PCLLYY320 pKa = 11.12YY321 pKa = 9.4ITIPNNGNKK330 pKa = 8.65SHH332 pKa = 7.52CIQALNLSGLLEE344 pKa = 4.05HH345 pKa = 7.19RR346 pKa = 11.84LLHH349 pKa = 6.25FGSLGTSCRR358 pKa = 11.84DD359 pKa = 3.15PSCRR363 pKa = 11.84VIGIDD368 pKa = 3.66RR369 pKa = 11.84PEE371 pKa = 4.75LYY373 pKa = 10.44SEE375 pKa = 4.69LNRR378 pKa = 11.84IVDD381 pKa = 3.37NTTIKK386 pKa = 10.82DD387 pKa = 3.33RR388 pKa = 11.84ADD390 pKa = 3.14AMLEE394 pKa = 3.87INADD398 pKa = 3.4EE399 pKa = 4.11MSSVKK404 pKa = 10.6SSGSGKK410 pKa = 10.43SLTKK414 pKa = 10.33GKK416 pKa = 9.48YY417 pKa = 8.57VRR419 pKa = 11.84NSIDD423 pKa = 3.06KK424 pKa = 10.4GHH426 pKa = 6.7NLTDD430 pKa = 3.63LGKK433 pKa = 10.71ANGDD437 pKa = 3.67QYY439 pKa = 11.77SHH441 pKa = 6.82YY442 pKa = 10.71SQMSKK447 pKa = 10.64PSGSEE452 pKa = 3.0ISKK455 pKa = 10.64FITAEE460 pKa = 3.88AVKK463 pKa = 10.44GISDD467 pKa = 3.78SSLDD471 pKa = 3.61ATEE474 pKa = 5.21LSLKK478 pKa = 10.57LAMLQSSAVTIGASKK493 pKa = 11.29NEE495 pKa = 4.14TKK497 pKa = 10.39PVEE500 pKa = 4.38TKK502 pKa = 10.04VVKK505 pKa = 10.55APSVNKK511 pKa = 9.05TADD514 pKa = 3.52EE515 pKa = 4.29LEE517 pKa = 4.58EE518 pKa = 4.05II519 pKa = 4.67
Molecular weight: 57.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6079 |
174 |
1222 |
506.6 |
56.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.284 ± 0.511 | 1.25 ± 0.241 |
6.317 ± 0.409 | 4.951 ± 0.196 |
3.866 ± 0.361 | 5.79 ± 0.272 |
2.5 ± 0.348 | 6.498 ± 0.383 |
5.083 ± 0.456 | 9.048 ± 0.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.158 ± 0.218 | 6.662 ± 0.296 |
3.915 ± 0.14 | 2.632 ± 0.186 |
5.593 ± 0.352 | 8.11 ± 0.446 |
6.021 ± 0.195 | 7.715 ± 0.339 |
0.592 ± 0.091 | 4.014 ± 0.368 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
