
Mycolicibacterium fallax (Mycobacterium fallax)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria;
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
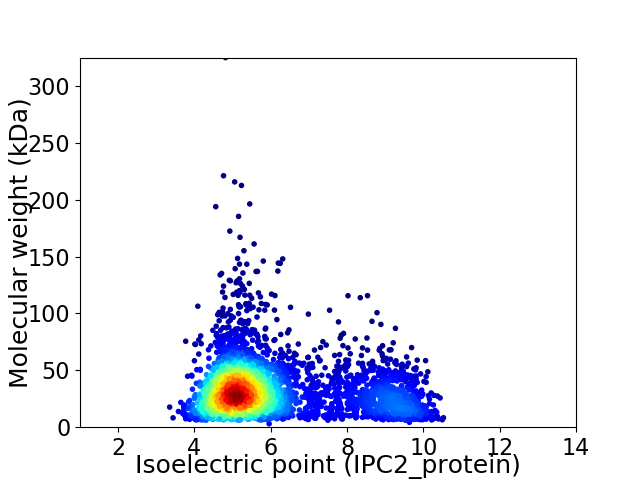
Virtual 2D-PAGE plot for 3787 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X1R7U7|A0A1X1R7U7_MYCFA Uncharacterized protein OS=Mycolicibacterium fallax OX=1793 GN=AWC04_15025 PE=4 SV=1
MM1 pKa = 7.94DD2 pKa = 5.11NDD4 pKa = 4.06IIALIDD10 pKa = 3.35QALDD14 pKa = 3.19EE15 pKa = 4.94GEE17 pKa = 4.69PYY19 pKa = 10.8SPADD23 pKa = 5.72DD24 pKa = 4.31PYY26 pKa = 10.28SWNDD30 pKa = 3.04AAVWTSTGDD39 pKa = 3.39HH40 pKa = 6.47QDD42 pKa = 3.13RR43 pKa = 11.84DD44 pKa = 3.65DD45 pKa = 5.06DD46 pKa = 4.01EE47 pKa = 7.21RR48 pKa = 11.84DD49 pKa = 3.82DD50 pKa = 4.36EE51 pKa = 4.65VDD53 pKa = 3.06PLRR56 pKa = 11.84YY57 pKa = 9.54VDD59 pKa = 4.8NPFAGTYY66 pKa = 8.95YY67 pKa = 10.2QDD69 pKa = 3.98PSDD72 pKa = 4.61PPWLVTPVGHH82 pKa = 5.47QHH84 pKa = 6.9IYY86 pKa = 11.09DD87 pKa = 3.83HH88 pKa = 7.61DD89 pKa = 4.17EE90 pKa = 4.0PAPAVWEE97 pKa = 4.49VSRR100 pKa = 11.84QPPGCYY106 pKa = 9.17SDD108 pKa = 5.01EE109 pKa = 4.35PQAGQTVVLSATLSDD124 pKa = 4.14GLRR127 pKa = 11.84TCYY130 pKa = 8.76FTYY133 pKa = 10.93VHH135 pKa = 6.97GEE137 pKa = 4.08DD138 pKa = 4.08PQHH141 pKa = 6.8GEE143 pKa = 3.57WDD145 pKa = 3.44GDD147 pKa = 3.5ADD149 pKa = 3.5MLEE152 pKa = 4.2AVRR155 pKa = 11.84ILGALDD161 pKa = 4.35PPDD164 pKa = 4.74DD165 pKa = 4.66PLRR168 pKa = 11.84MIAGALARR176 pKa = 11.84VGWWGTVSDD185 pKa = 4.81SVWQVLAGDD194 pKa = 4.95DD195 pKa = 3.88YY196 pKa = 12.2
MM1 pKa = 7.94DD2 pKa = 5.11NDD4 pKa = 4.06IIALIDD10 pKa = 3.35QALDD14 pKa = 3.19EE15 pKa = 4.94GEE17 pKa = 4.69PYY19 pKa = 10.8SPADD23 pKa = 5.72DD24 pKa = 4.31PYY26 pKa = 10.28SWNDD30 pKa = 3.04AAVWTSTGDD39 pKa = 3.39HH40 pKa = 6.47QDD42 pKa = 3.13RR43 pKa = 11.84DD44 pKa = 3.65DD45 pKa = 5.06DD46 pKa = 4.01EE47 pKa = 7.21RR48 pKa = 11.84DD49 pKa = 3.82DD50 pKa = 4.36EE51 pKa = 4.65VDD53 pKa = 3.06PLRR56 pKa = 11.84YY57 pKa = 9.54VDD59 pKa = 4.8NPFAGTYY66 pKa = 8.95YY67 pKa = 10.2QDD69 pKa = 3.98PSDD72 pKa = 4.61PPWLVTPVGHH82 pKa = 5.47QHH84 pKa = 6.9IYY86 pKa = 11.09DD87 pKa = 3.83HH88 pKa = 7.61DD89 pKa = 4.17EE90 pKa = 4.0PAPAVWEE97 pKa = 4.49VSRR100 pKa = 11.84QPPGCYY106 pKa = 9.17SDD108 pKa = 5.01EE109 pKa = 4.35PQAGQTVVLSATLSDD124 pKa = 4.14GLRR127 pKa = 11.84TCYY130 pKa = 8.76FTYY133 pKa = 10.93VHH135 pKa = 6.97GEE137 pKa = 4.08DD138 pKa = 4.08PQHH141 pKa = 6.8GEE143 pKa = 3.57WDD145 pKa = 3.44GDD147 pKa = 3.5ADD149 pKa = 3.5MLEE152 pKa = 4.2AVRR155 pKa = 11.84ILGALDD161 pKa = 4.35PPDD164 pKa = 4.74DD165 pKa = 4.66PLRR168 pKa = 11.84MIAGALARR176 pKa = 11.84VGWWGTVSDD185 pKa = 4.81SVWQVLAGDD194 pKa = 4.95DD195 pKa = 3.88YY196 pKa = 12.2
Molecular weight: 21.81 kDa
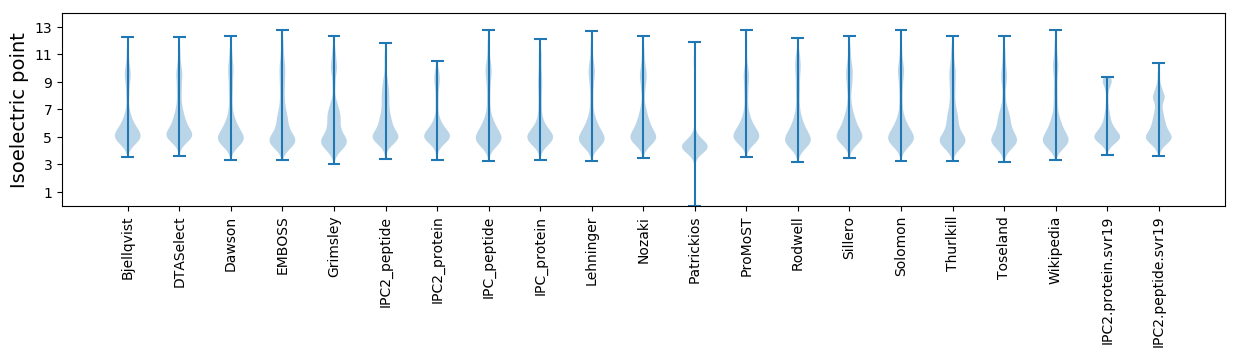
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X1RG22|A0A1X1RG22_MYCFA MarR family transcriptional regulator OS=Mycolicibacterium fallax OX=1793 GN=AWC04_07295 PE=4 SV=1
MM1 pKa = 7.03QRR3 pKa = 11.84RR4 pKa = 11.84NRR6 pKa = 11.84RR7 pKa = 11.84AGVEE11 pKa = 4.03DD12 pKa = 3.19RR13 pKa = 11.84WRR15 pKa = 11.84RR16 pKa = 11.84SDD18 pKa = 3.37GSPTARR24 pKa = 11.84SGMGRR29 pKa = 11.84RR30 pKa = 11.84WLARR34 pKa = 11.84YY35 pKa = 9.79VDD37 pKa = 3.94DD38 pKa = 5.11QGGEE42 pKa = 4.0NTRR45 pKa = 11.84SFDD48 pKa = 4.05RR49 pKa = 11.84KK50 pKa = 9.99VDD52 pKa = 3.37AQAFLNEE59 pKa = 3.63ITAAQTIGTYY69 pKa = 9.19VAPKK73 pKa = 9.75AGRR76 pKa = 11.84ITVRR80 pKa = 11.84EE81 pKa = 4.06LHH83 pKa = 6.31GKK85 pKa = 8.01WLGTQGHH92 pKa = 6.39LKK94 pKa = 8.21EE95 pKa = 4.48TTVATRR101 pKa = 11.84AFAWSGYY108 pKa = 9.27VEE110 pKa = 4.29GRR112 pKa = 11.84WAAVAVADD120 pKa = 4.31VQSSDD125 pKa = 2.39IRR127 pKa = 11.84AWVQQLAAGGAKK139 pKa = 9.81PATIEE144 pKa = 3.92NALSVLRR151 pKa = 11.84QILEE155 pKa = 3.92MAVDD159 pKa = 3.92DD160 pKa = 5.0RR161 pKa = 11.84RR162 pKa = 11.84IPRR165 pKa = 11.84NPCTGVKK172 pKa = 9.7SPRR175 pKa = 11.84RR176 pKa = 11.84QHH178 pKa = 6.43RR179 pKa = 11.84ARR181 pKa = 11.84GYY183 pKa = 8.21LTHH186 pKa = 6.17QQVEE190 pKa = 4.16LLARR194 pKa = 11.84EE195 pKa = 4.12VGEE198 pKa = 4.04YY199 pKa = 10.71AVVVRR204 pKa = 11.84FLAYY208 pKa = 9.27TGLRR212 pKa = 11.84WGEE215 pKa = 3.76MAALRR220 pKa = 11.84VEE222 pKa = 4.32SFDD225 pKa = 3.24MLRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84VNIRR234 pKa = 11.84EE235 pKa = 3.81AVAEE239 pKa = 4.26VKK241 pKa = 10.74GRR243 pKa = 11.84VVWSSPKK250 pKa = 8.22SHH252 pKa = 6.39EE253 pKa = 4.18RR254 pKa = 11.84RR255 pKa = 11.84SVPFPAFLADD265 pKa = 3.7PLAAIMIGKK274 pKa = 9.37RR275 pKa = 11.84RR276 pKa = 11.84DD277 pKa = 3.61DD278 pKa = 4.18LVFTSPGGALLRR290 pKa = 11.84VSTWRR295 pKa = 11.84PRR297 pKa = 11.84VFNMAVQRR305 pKa = 11.84LQEE308 pKa = 4.53ADD310 pKa = 3.23PAYY313 pKa = 8.32PTVTPHH319 pKa = 7.59DD320 pKa = 4.26LRR322 pKa = 11.84HH323 pKa = 5.48TAASLSISAGANVKK337 pKa = 9.86AVQTMLGHH345 pKa = 6.81ASAVLTLDD353 pKa = 3.76TYY355 pKa = 12.12ADD357 pKa = 4.59LFPDD361 pKa = 4.06DD362 pKa = 4.89LEE364 pKa = 4.43QVSVALDD371 pKa = 3.57AARR374 pKa = 11.84MRR376 pKa = 11.84SLAATADD383 pKa = 3.49QLRR386 pKa = 11.84TGKK389 pKa = 10.35
MM1 pKa = 7.03QRR3 pKa = 11.84RR4 pKa = 11.84NRR6 pKa = 11.84RR7 pKa = 11.84AGVEE11 pKa = 4.03DD12 pKa = 3.19RR13 pKa = 11.84WRR15 pKa = 11.84RR16 pKa = 11.84SDD18 pKa = 3.37GSPTARR24 pKa = 11.84SGMGRR29 pKa = 11.84RR30 pKa = 11.84WLARR34 pKa = 11.84YY35 pKa = 9.79VDD37 pKa = 3.94DD38 pKa = 5.11QGGEE42 pKa = 4.0NTRR45 pKa = 11.84SFDD48 pKa = 4.05RR49 pKa = 11.84KK50 pKa = 9.99VDD52 pKa = 3.37AQAFLNEE59 pKa = 3.63ITAAQTIGTYY69 pKa = 9.19VAPKK73 pKa = 9.75AGRR76 pKa = 11.84ITVRR80 pKa = 11.84EE81 pKa = 4.06LHH83 pKa = 6.31GKK85 pKa = 8.01WLGTQGHH92 pKa = 6.39LKK94 pKa = 8.21EE95 pKa = 4.48TTVATRR101 pKa = 11.84AFAWSGYY108 pKa = 9.27VEE110 pKa = 4.29GRR112 pKa = 11.84WAAVAVADD120 pKa = 4.31VQSSDD125 pKa = 2.39IRR127 pKa = 11.84AWVQQLAAGGAKK139 pKa = 9.81PATIEE144 pKa = 3.92NALSVLRR151 pKa = 11.84QILEE155 pKa = 3.92MAVDD159 pKa = 3.92DD160 pKa = 5.0RR161 pKa = 11.84RR162 pKa = 11.84IPRR165 pKa = 11.84NPCTGVKK172 pKa = 9.7SPRR175 pKa = 11.84RR176 pKa = 11.84QHH178 pKa = 6.43RR179 pKa = 11.84ARR181 pKa = 11.84GYY183 pKa = 8.21LTHH186 pKa = 6.17QQVEE190 pKa = 4.16LLARR194 pKa = 11.84EE195 pKa = 4.12VGEE198 pKa = 4.04YY199 pKa = 10.71AVVVRR204 pKa = 11.84FLAYY208 pKa = 9.27TGLRR212 pKa = 11.84WGEE215 pKa = 3.76MAALRR220 pKa = 11.84VEE222 pKa = 4.32SFDD225 pKa = 3.24MLRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84VNIRR234 pKa = 11.84EE235 pKa = 3.81AVAEE239 pKa = 4.26VKK241 pKa = 10.74GRR243 pKa = 11.84VVWSSPKK250 pKa = 8.22SHH252 pKa = 6.39EE253 pKa = 4.18RR254 pKa = 11.84RR255 pKa = 11.84SVPFPAFLADD265 pKa = 3.7PLAAIMIGKK274 pKa = 9.37RR275 pKa = 11.84RR276 pKa = 11.84DD277 pKa = 3.61DD278 pKa = 4.18LVFTSPGGALLRR290 pKa = 11.84VSTWRR295 pKa = 11.84PRR297 pKa = 11.84VFNMAVQRR305 pKa = 11.84LQEE308 pKa = 4.53ADD310 pKa = 3.23PAYY313 pKa = 8.32PTVTPHH319 pKa = 7.59DD320 pKa = 4.26LRR322 pKa = 11.84HH323 pKa = 5.48TAASLSISAGANVKK337 pKa = 9.86AVQTMLGHH345 pKa = 6.81ASAVLTLDD353 pKa = 3.76TYY355 pKa = 12.12ADD357 pKa = 4.59LFPDD361 pKa = 4.06DD362 pKa = 4.89LEE364 pKa = 4.43QVSVALDD371 pKa = 3.57AARR374 pKa = 11.84MRR376 pKa = 11.84SLAATADD383 pKa = 3.49QLRR386 pKa = 11.84TGKK389 pKa = 10.35
Molecular weight: 43.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1219274 |
29 |
3072 |
322.0 |
34.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.042 ± 0.057 | 0.777 ± 0.009 |
6.321 ± 0.033 | 5.193 ± 0.029 |
2.886 ± 0.022 | 9.183 ± 0.042 |
2.063 ± 0.021 | 4.147 ± 0.026 |
2.017 ± 0.029 | 10.068 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.974 ± 0.017 | 2.122 ± 0.019 |
6.058 ± 0.04 | 2.851 ± 0.021 |
7.452 ± 0.041 | 4.91 ± 0.027 |
5.955 ± 0.027 | 8.42 ± 0.034 |
1.472 ± 0.017 | 2.089 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
