
Actinospica robiniae DSM 44927
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Catenulisporales; Actinospicaceae; Actinospica; Actinospica robiniae
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
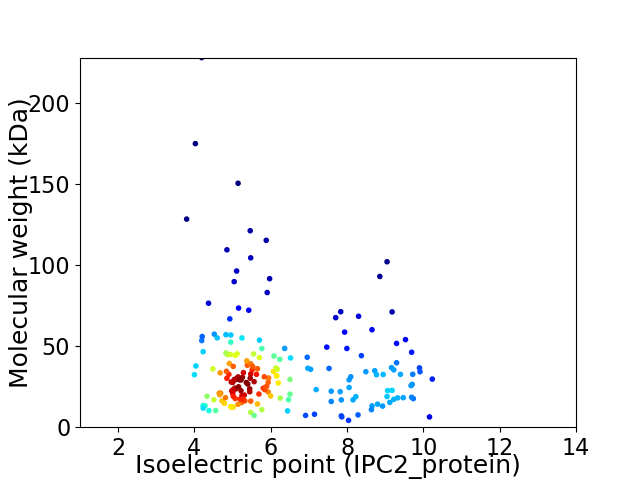
Virtual 2D-PAGE plot for 191 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W9DYU4|W9DYU4_9ACTN Acyl-CoA dehydrogenase OS=Actinospica robiniae DSM 44927 OX=479430 GN=ActroDRAFT_0011 PE=3 SV=1
MM1 pKa = 7.46RR2 pKa = 11.84RR3 pKa = 11.84VLTSVISILTATAALSPLGSAAQAADD29 pKa = 3.36ATAPPPLKK37 pKa = 10.24VVTFNTCGQSTACQSTEE54 pKa = 4.95DD55 pKa = 3.81LTDD58 pKa = 3.22WASDD62 pKa = 3.5LSSAILAYY70 pKa = 10.43GPDD73 pKa = 3.34AGSIQEE79 pKa = 4.06VCGNQVDD86 pKa = 4.25AFSADD91 pKa = 3.21LTGYY95 pKa = 7.71TVYY98 pKa = 10.63FNPAWDD104 pKa = 4.58LPTPATTGTNNNHH117 pKa = 6.02CSHH120 pKa = 7.16WASTEE125 pKa = 3.97NEE127 pKa = 4.29DD128 pKa = 4.49EE129 pKa = 4.73FGDD132 pKa = 3.84MIYY135 pKa = 10.75VKK137 pKa = 10.5SSEE140 pKa = 4.38SPVLSTDD147 pKa = 3.49VIYY150 pKa = 7.83PTNPTVEE157 pKa = 4.31SRR159 pKa = 11.84PLEE162 pKa = 4.34CVEE165 pKa = 4.24ATDD168 pKa = 4.77SGMPYY173 pKa = 9.89QYY175 pKa = 10.95CDD177 pKa = 3.07VHH179 pKa = 8.34LDD181 pKa = 3.54GGLGGAPAQGLPEE194 pKa = 4.51VINRR198 pKa = 11.84MQYY201 pKa = 9.2WGGANTPTVLAGDD214 pKa = 4.65FNADD218 pKa = 3.25PTDD221 pKa = 3.83PNMGLIYY228 pKa = 9.96QAQDD232 pKa = 3.1GNGSFIEE239 pKa = 4.12AEE241 pKa = 4.36QYY243 pKa = 11.04DD244 pKa = 4.02KK245 pKa = 11.46AYY247 pKa = 7.59FTPQCRR253 pKa = 11.84QLTSCRR259 pKa = 11.84SGAPTTYY266 pKa = 9.98PVLGSKK272 pKa = 11.14GEE274 pKa = 4.12VAAGGQARR282 pKa = 11.84KK283 pKa = 9.36FDD285 pKa = 4.62YY286 pKa = 10.85IFATAKK292 pKa = 10.62DD293 pKa = 4.47FSADD297 pKa = 2.97SDD299 pKa = 4.53SIVEE303 pKa = 4.01VKK305 pKa = 10.66AADD308 pKa = 4.02GTDD311 pKa = 3.48LTHH314 pKa = 7.03NDD316 pKa = 3.08HH317 pKa = 7.21LMYY320 pKa = 10.54QATFDD325 pKa = 3.39WRR327 pKa = 11.84AATTPTPAPAAPTVLTNPRR346 pKa = 11.84TIGSEE351 pKa = 4.16DD352 pKa = 3.02PWANAVDD359 pKa = 3.92YY360 pKa = 8.75TAGNFLGTGKK370 pKa = 10.79ADD372 pKa = 3.38MVARR376 pKa = 11.84YY377 pKa = 10.04ADD379 pKa = 4.15GTVMLYY385 pKa = 9.87PGKK388 pKa = 10.73GGGAFGSGQQLEE400 pKa = 4.68LATQYY405 pKa = 11.49GFYY408 pKa = 10.39AALGRR413 pKa = 11.84GVASVTAGDD422 pKa = 4.01FDD424 pKa = 5.83GDD426 pKa = 3.89GANDD430 pKa = 3.25GRR432 pKa = 11.84ADD434 pKa = 5.83LLVSWNNGDD443 pKa = 4.18VSLSPNNGSGGLGVSIPLLPGHH465 pKa = 6.18SSIFSKK471 pKa = 8.12MTSVGVGHH479 pKa = 6.33VLGSTDD485 pKa = 3.27TAVNDD490 pKa = 4.57AVFLTANRR498 pKa = 11.84SPIISLSPPPSSTAGPPTTASYY520 pKa = 11.36SVVAYY525 pKa = 9.29PIDD528 pKa = 3.91RR529 pKa = 11.84SEE531 pKa = 4.3SGTYY535 pKa = 9.76SAGSPVSLDD544 pKa = 3.35LGGATSLTDD553 pKa = 3.56VVDD556 pKa = 3.54MTVGDD561 pKa = 3.88FTGIGHH567 pKa = 7.4AEE569 pKa = 3.54ILLRR573 pKa = 11.84HH574 pKa = 6.0SDD576 pKa = 3.73GTLSLYY582 pKa = 10.89GDD584 pKa = 3.76NAQSDD589 pKa = 4.05FTAAPLTVVDD599 pKa = 3.88NRR601 pKa = 11.84NWAALTKK608 pKa = 10.49APTPSWQGVEE618 pKa = 4.0NLVAGDD624 pKa = 3.82FDD626 pKa = 4.16GTGKK630 pKa = 10.53DD631 pKa = 3.59GVVLQWMNEE640 pKa = 4.04YY641 pKa = 10.58QGTLVPGRR649 pKa = 11.84GQTEE653 pKa = 4.38YY654 pKa = 11.16LSSTGTAPGKK664 pKa = 10.33LFNAPVQITSGRR676 pKa = 11.84FEE678 pKa = 4.47PWTTSADD685 pKa = 3.37FAAATTTSSGDD696 pKa = 2.99TDD698 pKa = 5.71LITRR702 pKa = 11.84VQGTPAVGDD711 pKa = 3.79NPASAGQTDD720 pKa = 4.19LWTGRR725 pKa = 11.84GTGTFGEE732 pKa = 4.59TEE734 pKa = 3.73IDD736 pKa = 3.63PAVEE740 pKa = 3.79PGYY743 pKa = 10.58LDD745 pKa = 3.02WATAVTVGGFGTSGAQEE762 pKa = 4.1LFAYY766 pKa = 8.73NASGGGVYY774 pKa = 10.57ALNAGTSEE782 pKa = 4.37GPPTTTVAGLQSVDD796 pKa = 2.76WANVVEE802 pKa = 4.42VVPGTFYY809 pKa = 11.1AGPYY813 pKa = 7.98SDD815 pKa = 5.32LVIRR819 pKa = 11.84YY820 pKa = 9.36ADD822 pKa = 3.49GSVQLFQNITGQNGTSPSVPTFAAPTQVEE851 pKa = 4.45PTGSDD856 pKa = 2.64GWGDD860 pKa = 3.73AEE862 pKa = 4.81QISAGDD868 pKa = 3.57FNGDD872 pKa = 3.28GNEE875 pKa = 4.04DD876 pKa = 3.85LLVRR880 pKa = 11.84GSAGSLLLYY889 pKa = 10.08PGDD892 pKa = 4.01GAGHH896 pKa = 6.8FGGSVVIDD904 pKa = 4.06SAPAMADD911 pKa = 3.36LVSVEE916 pKa = 3.86QGTVYY921 pKa = 9.22GTAAGARR928 pKa = 11.84NIYY931 pKa = 10.02RR932 pKa = 11.84FSDD935 pKa = 3.66GEE937 pKa = 4.19AIIDD941 pKa = 3.72NGSTNLVDD949 pKa = 5.11PGTQSVNGTSASFLFHH965 pKa = 7.47SDD967 pKa = 3.74DD968 pKa = 3.65DD969 pKa = 4.07TGAIQSFSYY978 pKa = 10.69QLDD981 pKa = 3.55GAPAVSLDD989 pKa = 3.46AFHH992 pKa = 7.92DD993 pKa = 4.03EE994 pKa = 4.63VTVSLQDD1001 pKa = 3.58LTPGSHH1007 pKa = 6.33TLTVTATNSADD1018 pKa = 3.43VTSAATTYY1026 pKa = 11.23GFTDD1030 pKa = 3.94DD1031 pKa = 4.5QNTGPIHH1038 pKa = 7.05AWPLSNGTGIDD1049 pKa = 3.66TVGGADD1055 pKa = 3.86LTLSSAGTSWTTDD1068 pKa = 3.37PTHH1071 pKa = 5.88GTVLTLDD1078 pKa = 3.6GTSGYY1083 pKa = 10.52GATSGPLIDD1092 pKa = 3.78TTGSFSISAWAEE1104 pKa = 3.76PAVSSAGVIASQDD1117 pKa = 3.31GTNISGFVLYY1127 pKa = 10.84ANAGGGWTFAMAPTDD1142 pKa = 3.92SATWTADD1149 pKa = 3.54RR1150 pKa = 11.84ASTANTTDD1158 pKa = 3.77NPGSWTYY1165 pKa = 9.94LTATYY1170 pKa = 10.63NAATDD1175 pKa = 3.65TMSLYY1180 pKa = 11.33VNGAPAATGTHH1191 pKa = 4.63TTTWNATGSFIIGRR1205 pKa = 11.84DD1206 pKa = 3.82FVDD1209 pKa = 3.52SAPNGFFNGDD1219 pKa = 3.13ISNVQAFNYY1228 pKa = 8.56PLSASQVTALYY1239 pKa = 10.06DD1240 pKa = 3.48QNTPPNQQ1247 pKa = 3.26
MM1 pKa = 7.46RR2 pKa = 11.84RR3 pKa = 11.84VLTSVISILTATAALSPLGSAAQAADD29 pKa = 3.36ATAPPPLKK37 pKa = 10.24VVTFNTCGQSTACQSTEE54 pKa = 4.95DD55 pKa = 3.81LTDD58 pKa = 3.22WASDD62 pKa = 3.5LSSAILAYY70 pKa = 10.43GPDD73 pKa = 3.34AGSIQEE79 pKa = 4.06VCGNQVDD86 pKa = 4.25AFSADD91 pKa = 3.21LTGYY95 pKa = 7.71TVYY98 pKa = 10.63FNPAWDD104 pKa = 4.58LPTPATTGTNNNHH117 pKa = 6.02CSHH120 pKa = 7.16WASTEE125 pKa = 3.97NEE127 pKa = 4.29DD128 pKa = 4.49EE129 pKa = 4.73FGDD132 pKa = 3.84MIYY135 pKa = 10.75VKK137 pKa = 10.5SSEE140 pKa = 4.38SPVLSTDD147 pKa = 3.49VIYY150 pKa = 7.83PTNPTVEE157 pKa = 4.31SRR159 pKa = 11.84PLEE162 pKa = 4.34CVEE165 pKa = 4.24ATDD168 pKa = 4.77SGMPYY173 pKa = 9.89QYY175 pKa = 10.95CDD177 pKa = 3.07VHH179 pKa = 8.34LDD181 pKa = 3.54GGLGGAPAQGLPEE194 pKa = 4.51VINRR198 pKa = 11.84MQYY201 pKa = 9.2WGGANTPTVLAGDD214 pKa = 4.65FNADD218 pKa = 3.25PTDD221 pKa = 3.83PNMGLIYY228 pKa = 9.96QAQDD232 pKa = 3.1GNGSFIEE239 pKa = 4.12AEE241 pKa = 4.36QYY243 pKa = 11.04DD244 pKa = 4.02KK245 pKa = 11.46AYY247 pKa = 7.59FTPQCRR253 pKa = 11.84QLTSCRR259 pKa = 11.84SGAPTTYY266 pKa = 9.98PVLGSKK272 pKa = 11.14GEE274 pKa = 4.12VAAGGQARR282 pKa = 11.84KK283 pKa = 9.36FDD285 pKa = 4.62YY286 pKa = 10.85IFATAKK292 pKa = 10.62DD293 pKa = 4.47FSADD297 pKa = 2.97SDD299 pKa = 4.53SIVEE303 pKa = 4.01VKK305 pKa = 10.66AADD308 pKa = 4.02GTDD311 pKa = 3.48LTHH314 pKa = 7.03NDD316 pKa = 3.08HH317 pKa = 7.21LMYY320 pKa = 10.54QATFDD325 pKa = 3.39WRR327 pKa = 11.84AATTPTPAPAAPTVLTNPRR346 pKa = 11.84TIGSEE351 pKa = 4.16DD352 pKa = 3.02PWANAVDD359 pKa = 3.92YY360 pKa = 8.75TAGNFLGTGKK370 pKa = 10.79ADD372 pKa = 3.38MVARR376 pKa = 11.84YY377 pKa = 10.04ADD379 pKa = 4.15GTVMLYY385 pKa = 9.87PGKK388 pKa = 10.73GGGAFGSGQQLEE400 pKa = 4.68LATQYY405 pKa = 11.49GFYY408 pKa = 10.39AALGRR413 pKa = 11.84GVASVTAGDD422 pKa = 4.01FDD424 pKa = 5.83GDD426 pKa = 3.89GANDD430 pKa = 3.25GRR432 pKa = 11.84ADD434 pKa = 5.83LLVSWNNGDD443 pKa = 4.18VSLSPNNGSGGLGVSIPLLPGHH465 pKa = 6.18SSIFSKK471 pKa = 8.12MTSVGVGHH479 pKa = 6.33VLGSTDD485 pKa = 3.27TAVNDD490 pKa = 4.57AVFLTANRR498 pKa = 11.84SPIISLSPPPSSTAGPPTTASYY520 pKa = 11.36SVVAYY525 pKa = 9.29PIDD528 pKa = 3.91RR529 pKa = 11.84SEE531 pKa = 4.3SGTYY535 pKa = 9.76SAGSPVSLDD544 pKa = 3.35LGGATSLTDD553 pKa = 3.56VVDD556 pKa = 3.54MTVGDD561 pKa = 3.88FTGIGHH567 pKa = 7.4AEE569 pKa = 3.54ILLRR573 pKa = 11.84HH574 pKa = 6.0SDD576 pKa = 3.73GTLSLYY582 pKa = 10.89GDD584 pKa = 3.76NAQSDD589 pKa = 4.05FTAAPLTVVDD599 pKa = 3.88NRR601 pKa = 11.84NWAALTKK608 pKa = 10.49APTPSWQGVEE618 pKa = 4.0NLVAGDD624 pKa = 3.82FDD626 pKa = 4.16GTGKK630 pKa = 10.53DD631 pKa = 3.59GVVLQWMNEE640 pKa = 4.04YY641 pKa = 10.58QGTLVPGRR649 pKa = 11.84GQTEE653 pKa = 4.38YY654 pKa = 11.16LSSTGTAPGKK664 pKa = 10.33LFNAPVQITSGRR676 pKa = 11.84FEE678 pKa = 4.47PWTTSADD685 pKa = 3.37FAAATTTSSGDD696 pKa = 2.99TDD698 pKa = 5.71LITRR702 pKa = 11.84VQGTPAVGDD711 pKa = 3.79NPASAGQTDD720 pKa = 4.19LWTGRR725 pKa = 11.84GTGTFGEE732 pKa = 4.59TEE734 pKa = 3.73IDD736 pKa = 3.63PAVEE740 pKa = 3.79PGYY743 pKa = 10.58LDD745 pKa = 3.02WATAVTVGGFGTSGAQEE762 pKa = 4.1LFAYY766 pKa = 8.73NASGGGVYY774 pKa = 10.57ALNAGTSEE782 pKa = 4.37GPPTTTVAGLQSVDD796 pKa = 2.76WANVVEE802 pKa = 4.42VVPGTFYY809 pKa = 11.1AGPYY813 pKa = 7.98SDD815 pKa = 5.32LVIRR819 pKa = 11.84YY820 pKa = 9.36ADD822 pKa = 3.49GSVQLFQNITGQNGTSPSVPTFAAPTQVEE851 pKa = 4.45PTGSDD856 pKa = 2.64GWGDD860 pKa = 3.73AEE862 pKa = 4.81QISAGDD868 pKa = 3.57FNGDD872 pKa = 3.28GNEE875 pKa = 4.04DD876 pKa = 3.85LLVRR880 pKa = 11.84GSAGSLLLYY889 pKa = 10.08PGDD892 pKa = 4.01GAGHH896 pKa = 6.8FGGSVVIDD904 pKa = 4.06SAPAMADD911 pKa = 3.36LVSVEE916 pKa = 3.86QGTVYY921 pKa = 9.22GTAAGARR928 pKa = 11.84NIYY931 pKa = 10.02RR932 pKa = 11.84FSDD935 pKa = 3.66GEE937 pKa = 4.19AIIDD941 pKa = 3.72NGSTNLVDD949 pKa = 5.11PGTQSVNGTSASFLFHH965 pKa = 7.47SDD967 pKa = 3.74DD968 pKa = 3.65DD969 pKa = 4.07TGAIQSFSYY978 pKa = 10.69QLDD981 pKa = 3.55GAPAVSLDD989 pKa = 3.46AFHH992 pKa = 7.92DD993 pKa = 4.03EE994 pKa = 4.63VTVSLQDD1001 pKa = 3.58LTPGSHH1007 pKa = 6.33TLTVTATNSADD1018 pKa = 3.43VTSAATTYY1026 pKa = 11.23GFTDD1030 pKa = 3.94DD1031 pKa = 4.5QNTGPIHH1038 pKa = 7.05AWPLSNGTGIDD1049 pKa = 3.66TVGGADD1055 pKa = 3.86LTLSSAGTSWTTDD1068 pKa = 3.37PTHH1071 pKa = 5.88GTVLTLDD1078 pKa = 3.6GTSGYY1083 pKa = 10.52GATSGPLIDD1092 pKa = 3.78TTGSFSISAWAEE1104 pKa = 3.76PAVSSAGVIASQDD1117 pKa = 3.31GTNISGFVLYY1127 pKa = 10.84ANAGGGWTFAMAPTDD1142 pKa = 3.92SATWTADD1149 pKa = 3.54RR1150 pKa = 11.84ASTANTTDD1158 pKa = 3.77NPGSWTYY1165 pKa = 9.94LTATYY1170 pKa = 10.63NAATDD1175 pKa = 3.65TMSLYY1180 pKa = 11.33VNGAPAATGTHH1191 pKa = 4.63TTTWNATGSFIIGRR1205 pKa = 11.84DD1206 pKa = 3.82FVDD1209 pKa = 3.52SAPNGFFNGDD1219 pKa = 3.13ISNVQAFNYY1228 pKa = 8.56PLSASQVTALYY1239 pKa = 10.06DD1240 pKa = 3.48QNTPPNQQ1247 pKa = 3.26
Molecular weight: 128.38 kDa
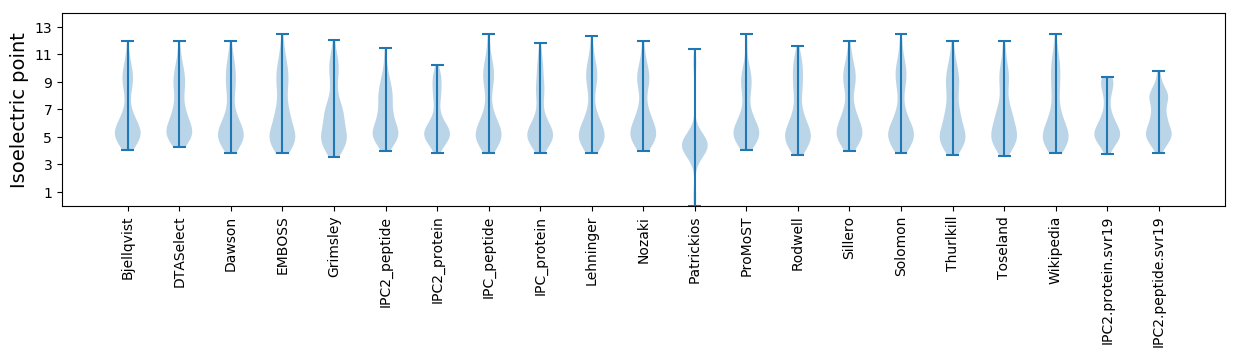
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W9DWG6|W9DWG6_9ACTN LamGL domain-containing protein OS=Actinospica robiniae DSM 44927 OX=479430 GN=ActroDRAFT_0206 PE=4 SV=1
MM1 pKa = 7.87ANRR4 pKa = 11.84PAAVKK9 pKa = 10.3SALKK13 pKa = 10.14SQVIEE18 pKa = 4.34TPSWGYY24 pKa = 11.01GNSGTRR30 pKa = 11.84FRR32 pKa = 11.84VFPQAGVPRR41 pKa = 11.84DD42 pKa = 3.89PFEE45 pKa = 4.9KK46 pKa = 10.53LADD49 pKa = 3.65AAAVHH54 pKa = 6.37AVTGVAAPVSLHH66 pKa = 5.85IPRR69 pKa = 11.84DD70 pKa = 3.29RR71 pKa = 11.84VADD74 pKa = 3.72YY75 pKa = 11.26GKK77 pKa = 10.56LADD80 pKa = 3.91HH81 pKa = 6.84AAGLGVRR88 pKa = 11.84VGMINSNTFQDD99 pKa = 4.06EE100 pKa = 4.14DD101 pKa = 4.12CKK103 pKa = 10.96LGSVCHH109 pKa = 6.64PDD111 pKa = 2.81VGVRR115 pKa = 11.84RR116 pKa = 11.84KK117 pKa = 7.89VTPASANGPRR127 pKa = 11.84PRR129 pKa = 11.84LGSQTGIEE137 pKa = 4.0PQLRR141 pKa = 11.84RR142 pKa = 11.84DD143 pKa = 3.79KK144 pKa = 11.31NAGSHH149 pKa = 6.38PGCHH153 pKa = 5.65GADD156 pKa = 3.16RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84AALHH163 pKa = 5.12VPAWSRR169 pKa = 11.84RR170 pKa = 11.84SS171 pKa = 3.32
MM1 pKa = 7.87ANRR4 pKa = 11.84PAAVKK9 pKa = 10.3SALKK13 pKa = 10.14SQVIEE18 pKa = 4.34TPSWGYY24 pKa = 11.01GNSGTRR30 pKa = 11.84FRR32 pKa = 11.84VFPQAGVPRR41 pKa = 11.84DD42 pKa = 3.89PFEE45 pKa = 4.9KK46 pKa = 10.53LADD49 pKa = 3.65AAAVHH54 pKa = 6.37AVTGVAAPVSLHH66 pKa = 5.85IPRR69 pKa = 11.84DD70 pKa = 3.29RR71 pKa = 11.84VADD74 pKa = 3.72YY75 pKa = 11.26GKK77 pKa = 10.56LADD80 pKa = 3.91HH81 pKa = 6.84AAGLGVRR88 pKa = 11.84VGMINSNTFQDD99 pKa = 4.06EE100 pKa = 4.14DD101 pKa = 4.12CKK103 pKa = 10.96LGSVCHH109 pKa = 6.64PDD111 pKa = 2.81VGVRR115 pKa = 11.84RR116 pKa = 11.84KK117 pKa = 7.89VTPASANGPRR127 pKa = 11.84PRR129 pKa = 11.84LGSQTGIEE137 pKa = 4.0PQLRR141 pKa = 11.84RR142 pKa = 11.84DD143 pKa = 3.79KK144 pKa = 11.31NAGSHH149 pKa = 6.38PGCHH153 pKa = 5.65GADD156 pKa = 3.16RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84AALHH163 pKa = 5.12VPAWSRR169 pKa = 11.84RR170 pKa = 11.84SS171 pKa = 3.32
Molecular weight: 18.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
66481 |
39 |
2170 |
348.1 |
37.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.379 ± 0.253 | 1.012 ± 0.05 |
5.614 ± 0.12 | 5.027 ± 0.201 |
2.736 ± 0.097 | 9.352 ± 0.16 |
2.156 ± 0.091 | 3.571 ± 0.101 |
1.602 ± 0.086 | 10.132 ± 0.258 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.641 ± 0.064 | 2.127 ± 0.164 |
5.761 ± 0.118 | 3.066 ± 0.1 |
7.387 ± 0.34 | 6.121 ± 0.227 |
6.516 ± 0.388 | 7.753 ± 0.152 |
1.652 ± 0.063 | 2.398 ± 0.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
