
Sewage-associated circular DNA virus-23
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.63
Get precalculated fractions of proteins
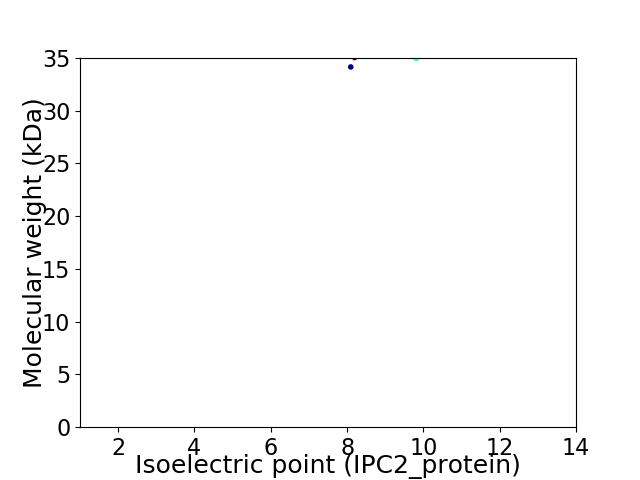
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH82|A0A0B4UH82_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-23 OX=1592090 PE=3 SV=1
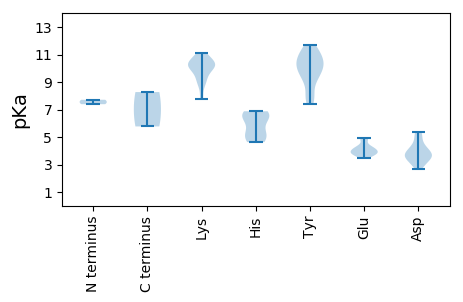
MM1 pKa = 7.71TSRR4 pKa = 11.84KK5 pKa = 7.75WVFTVNNYY13 pKa = 9.04TDD15 pKa = 5.17DD16 pKa = 3.29EE17 pKa = 4.48WNEE20 pKa = 3.78LKK22 pKa = 10.33RR23 pKa = 11.84WCMVNTKK30 pKa = 9.52YY31 pKa = 10.99AIMGKK36 pKa = 9.13EE37 pKa = 3.98VGPQCGTPHH46 pKa = 5.4IQGYY50 pKa = 10.2LSANKK55 pKa = 8.29PHH57 pKa = 6.8RR58 pKa = 11.84RR59 pKa = 11.84SGMVEE64 pKa = 3.5VCRR67 pKa = 11.84RR68 pKa = 11.84ARR70 pKa = 11.84WEE72 pKa = 3.74AAKK75 pKa = 11.14GNDD78 pKa = 3.7TQNDD82 pKa = 4.2DD83 pKa = 4.13YY84 pKa = 11.32CRR86 pKa = 11.84KK87 pKa = 9.16AAKK90 pKa = 9.72EE91 pKa = 3.75GRR93 pKa = 11.84EE94 pKa = 4.27VWRR97 pKa = 11.84HH98 pKa = 4.63GQPCTGAGQRR108 pKa = 11.84TDD110 pKa = 3.79LLRR113 pKa = 11.84VAAAVDD119 pKa = 3.55SGVSLRR125 pKa = 11.84EE126 pKa = 3.75IASEE130 pKa = 4.0NPSEE134 pKa = 4.27FIKK137 pKa = 9.92FHH139 pKa = 6.63RR140 pKa = 11.84GILAYY145 pKa = 10.41RR146 pKa = 11.84NLVRR150 pKa = 11.84PAVPRR155 pKa = 11.84DD156 pKa = 3.78FKK158 pKa = 11.11TEE160 pKa = 3.51LHH162 pKa = 6.19VFVGPPGSGKK172 pKa = 8.75SRR174 pKa = 11.84RR175 pKa = 11.84AQEE178 pKa = 4.03LASSFGTVYY187 pKa = 10.57RR188 pKa = 11.84KK189 pKa = 10.03VRR191 pKa = 11.84GPWWNGYY198 pKa = 7.37EE199 pKa = 4.06QQQSVIIDD207 pKa = 3.88DD208 pKa = 4.55FYY210 pKa = 11.41GWVPFDD216 pKa = 3.47EE217 pKa = 4.9LLRR220 pKa = 11.84VADD223 pKa = 5.18RR224 pKa = 11.84YY225 pKa = 7.84QHH227 pKa = 4.75QVEE230 pKa = 4.22IKK232 pKa = 10.67GGFEE236 pKa = 4.01EE237 pKa = 4.88FNSKK241 pKa = 10.32IIIITSNRR249 pKa = 11.84PIKK252 pKa = 10.28DD253 pKa = 2.71WYY255 pKa = 10.85KK256 pKa = 10.13FDD258 pKa = 4.69SYY260 pKa = 11.67DD261 pKa = 3.38PEE263 pKa = 4.8ALYY266 pKa = 10.49RR267 pKa = 11.84RR268 pKa = 11.84CSVYY272 pKa = 9.42QHH274 pKa = 6.51INMDD278 pKa = 3.65HH279 pKa = 6.51SVTDD283 pKa = 3.61YY284 pKa = 11.47EE285 pKa = 4.05IMEE288 pKa = 4.94GIKK291 pKa = 10.11INYY294 pKa = 8.27
MM1 pKa = 7.71TSRR4 pKa = 11.84KK5 pKa = 7.75WVFTVNNYY13 pKa = 9.04TDD15 pKa = 5.17DD16 pKa = 3.29EE17 pKa = 4.48WNEE20 pKa = 3.78LKK22 pKa = 10.33RR23 pKa = 11.84WCMVNTKK30 pKa = 9.52YY31 pKa = 10.99AIMGKK36 pKa = 9.13EE37 pKa = 3.98VGPQCGTPHH46 pKa = 5.4IQGYY50 pKa = 10.2LSANKK55 pKa = 8.29PHH57 pKa = 6.8RR58 pKa = 11.84RR59 pKa = 11.84SGMVEE64 pKa = 3.5VCRR67 pKa = 11.84RR68 pKa = 11.84ARR70 pKa = 11.84WEE72 pKa = 3.74AAKK75 pKa = 11.14GNDD78 pKa = 3.7TQNDD82 pKa = 4.2DD83 pKa = 4.13YY84 pKa = 11.32CRR86 pKa = 11.84KK87 pKa = 9.16AAKK90 pKa = 9.72EE91 pKa = 3.75GRR93 pKa = 11.84EE94 pKa = 4.27VWRR97 pKa = 11.84HH98 pKa = 4.63GQPCTGAGQRR108 pKa = 11.84TDD110 pKa = 3.79LLRR113 pKa = 11.84VAAAVDD119 pKa = 3.55SGVSLRR125 pKa = 11.84EE126 pKa = 3.75IASEE130 pKa = 4.0NPSEE134 pKa = 4.27FIKK137 pKa = 9.92FHH139 pKa = 6.63RR140 pKa = 11.84GILAYY145 pKa = 10.41RR146 pKa = 11.84NLVRR150 pKa = 11.84PAVPRR155 pKa = 11.84DD156 pKa = 3.78FKK158 pKa = 11.11TEE160 pKa = 3.51LHH162 pKa = 6.19VFVGPPGSGKK172 pKa = 8.75SRR174 pKa = 11.84RR175 pKa = 11.84AQEE178 pKa = 4.03LASSFGTVYY187 pKa = 10.57RR188 pKa = 11.84KK189 pKa = 10.03VRR191 pKa = 11.84GPWWNGYY198 pKa = 7.37EE199 pKa = 4.06QQQSVIIDD207 pKa = 3.88DD208 pKa = 4.55FYY210 pKa = 11.41GWVPFDD216 pKa = 3.47EE217 pKa = 4.9LLRR220 pKa = 11.84VADD223 pKa = 5.18RR224 pKa = 11.84YY225 pKa = 7.84QHH227 pKa = 4.75QVEE230 pKa = 4.22IKK232 pKa = 10.67GGFEE236 pKa = 4.01EE237 pKa = 4.88FNSKK241 pKa = 10.32IIIITSNRR249 pKa = 11.84PIKK252 pKa = 10.28DD253 pKa = 2.71WYY255 pKa = 10.85KK256 pKa = 10.13FDD258 pKa = 4.69SYY260 pKa = 11.67DD261 pKa = 3.38PEE263 pKa = 4.8ALYY266 pKa = 10.49RR267 pKa = 11.84RR268 pKa = 11.84CSVYY272 pKa = 9.42QHH274 pKa = 6.51INMDD278 pKa = 3.65HH279 pKa = 6.51SVTDD283 pKa = 3.61YY284 pKa = 11.47EE285 pKa = 4.05IMEE288 pKa = 4.94GIKK291 pKa = 10.11INYY294 pKa = 8.27
Molecular weight: 34.13 kDa
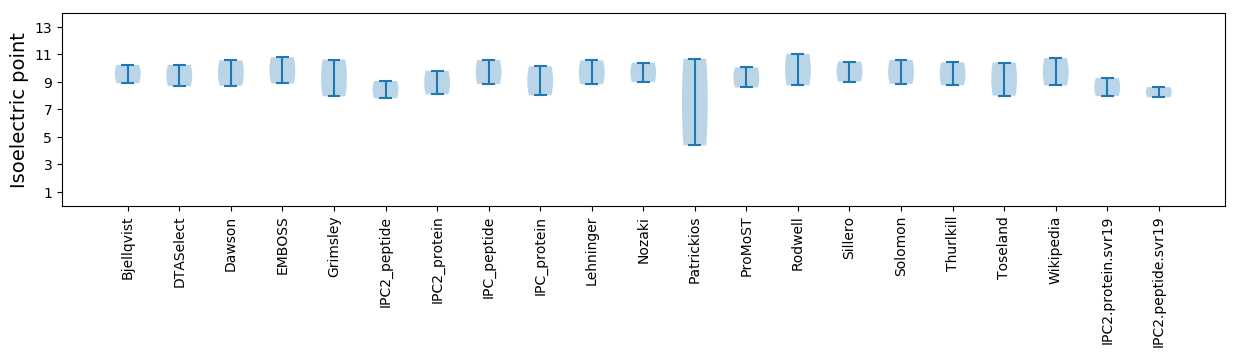
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UH82|A0A0B4UH82_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-23 OX=1592090 PE=3 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84YY4 pKa = 8.27PKK6 pKa = 10.7ASGNRR11 pKa = 11.84YY12 pKa = 9.27APYY15 pKa = 9.93IRR17 pKa = 11.84TAGRR21 pKa = 11.84MYY23 pKa = 10.83GAAKK27 pKa = 9.6KK28 pKa = 10.53AYY30 pKa = 9.45GIYY33 pKa = 9.98SKK35 pKa = 10.99VRR37 pKa = 11.84TRR39 pKa = 11.84YY40 pKa = 9.07RR41 pKa = 11.84NSGSRR46 pKa = 11.84TKK48 pKa = 10.49QKK50 pKa = 10.4RR51 pKa = 11.84NYY53 pKa = 7.86RR54 pKa = 11.84KK55 pKa = 10.04KK56 pKa = 10.12YY57 pKa = 7.76GGAMGSSIPGRR68 pKa = 11.84CGIYY72 pKa = 9.64VRR74 pKa = 11.84KK75 pKa = 9.57GRR77 pKa = 11.84KK78 pKa = 8.99DD79 pKa = 2.92RR80 pKa = 11.84WLKK83 pKa = 10.87NLTKK87 pKa = 10.59NSPPRR92 pKa = 11.84LLLNQSTNQVEE103 pKa = 4.23SDD105 pKa = 3.15AGKK108 pKa = 10.38QGTLSILQTLTRR120 pKa = 11.84TDD122 pKa = 4.12FMNMKK127 pKa = 8.87ATLPAISDD135 pKa = 3.52TGLGSGAAVGFATVSYY151 pKa = 11.19YY152 pKa = 10.17MDD154 pKa = 3.79IALKK158 pKa = 10.19RR159 pKa = 11.84LKK161 pKa = 10.9AEE163 pKa = 3.7ILITNTCNSVVFVKK177 pKa = 10.07MRR179 pKa = 11.84TWLCKK184 pKa = 9.9KK185 pKa = 10.26SSNASPLEE193 pKa = 3.75KK194 pKa = 9.42WGEE197 pKa = 3.82ILNVQNANVIPANKK211 pKa = 9.79VLTTTYY217 pKa = 10.71GATPYY222 pKa = 10.08MVDD225 pKa = 5.39GFNTWWKK232 pKa = 10.31CVQTRR237 pKa = 11.84NLRR240 pKa = 11.84LAPGEE245 pKa = 4.3IYY247 pKa = 10.61CHH249 pKa = 5.05NHH251 pKa = 5.24IDD253 pKa = 4.23KK254 pKa = 10.41INKK257 pKa = 8.96IIRR260 pKa = 11.84GSSYY264 pKa = 9.93EE265 pKa = 4.0AEE267 pKa = 3.92QNLYY271 pKa = 10.03IRR273 pKa = 11.84GLSKK277 pKa = 10.78VVTITIHH284 pKa = 6.88GEE286 pKa = 3.72PTQDD290 pKa = 3.04STGVNTLSGAVVNVAGKK307 pKa = 10.27RR308 pKa = 11.84KK309 pKa = 7.74PTQIHH314 pKa = 5.77
MM1 pKa = 7.39ARR3 pKa = 11.84YY4 pKa = 8.27PKK6 pKa = 10.7ASGNRR11 pKa = 11.84YY12 pKa = 9.27APYY15 pKa = 9.93IRR17 pKa = 11.84TAGRR21 pKa = 11.84MYY23 pKa = 10.83GAAKK27 pKa = 9.6KK28 pKa = 10.53AYY30 pKa = 9.45GIYY33 pKa = 9.98SKK35 pKa = 10.99VRR37 pKa = 11.84TRR39 pKa = 11.84YY40 pKa = 9.07RR41 pKa = 11.84NSGSRR46 pKa = 11.84TKK48 pKa = 10.49QKK50 pKa = 10.4RR51 pKa = 11.84NYY53 pKa = 7.86RR54 pKa = 11.84KK55 pKa = 10.04KK56 pKa = 10.12YY57 pKa = 7.76GGAMGSSIPGRR68 pKa = 11.84CGIYY72 pKa = 9.64VRR74 pKa = 11.84KK75 pKa = 9.57GRR77 pKa = 11.84KK78 pKa = 8.99DD79 pKa = 2.92RR80 pKa = 11.84WLKK83 pKa = 10.87NLTKK87 pKa = 10.59NSPPRR92 pKa = 11.84LLLNQSTNQVEE103 pKa = 4.23SDD105 pKa = 3.15AGKK108 pKa = 10.38QGTLSILQTLTRR120 pKa = 11.84TDD122 pKa = 4.12FMNMKK127 pKa = 8.87ATLPAISDD135 pKa = 3.52TGLGSGAAVGFATVSYY151 pKa = 11.19YY152 pKa = 10.17MDD154 pKa = 3.79IALKK158 pKa = 10.19RR159 pKa = 11.84LKK161 pKa = 10.9AEE163 pKa = 3.7ILITNTCNSVVFVKK177 pKa = 10.07MRR179 pKa = 11.84TWLCKK184 pKa = 9.9KK185 pKa = 10.26SSNASPLEE193 pKa = 3.75KK194 pKa = 9.42WGEE197 pKa = 3.82ILNVQNANVIPANKK211 pKa = 9.79VLTTTYY217 pKa = 10.71GATPYY222 pKa = 10.08MVDD225 pKa = 5.39GFNTWWKK232 pKa = 10.31CVQTRR237 pKa = 11.84NLRR240 pKa = 11.84LAPGEE245 pKa = 4.3IYY247 pKa = 10.61CHH249 pKa = 5.05NHH251 pKa = 5.24IDD253 pKa = 4.23KK254 pKa = 10.41INKK257 pKa = 8.96IIRR260 pKa = 11.84GSSYY264 pKa = 9.93EE265 pKa = 4.0AEE267 pKa = 3.92QNLYY271 pKa = 10.03IRR273 pKa = 11.84GLSKK277 pKa = 10.78VVTITIHH284 pKa = 6.88GEE286 pKa = 3.72PTQDD290 pKa = 3.04STGVNTLSGAVVNVAGKK307 pKa = 10.27RR308 pKa = 11.84KK309 pKa = 7.74PTQIHH314 pKa = 5.77
Molecular weight: 34.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
608 |
294 |
314 |
304.0 |
34.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.908 ± 0.549 | 1.809 ± 0.162 |
4.112 ± 1.167 | 4.605 ± 1.535 |
2.467 ± 0.89 | 8.059 ± 0.402 |
1.974 ± 0.522 | 6.086 ± 0.212 |
7.237 ± 1.016 | 5.592 ± 1.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.303 ± 0.183 | 5.921 ± 0.81 |
4.112 ± 0.216 | 3.618 ± 0.324 |
8.059 ± 0.548 | 6.414 ± 0.442 |
6.414 ± 1.629 | 6.743 ± 0.517 |
2.303 ± 0.53 | 5.263 ± 0.113 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
