
Skermanella stibiiresistens SB22
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Azospirillaceae; Skermanella; Skermanella stibiiresistens
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
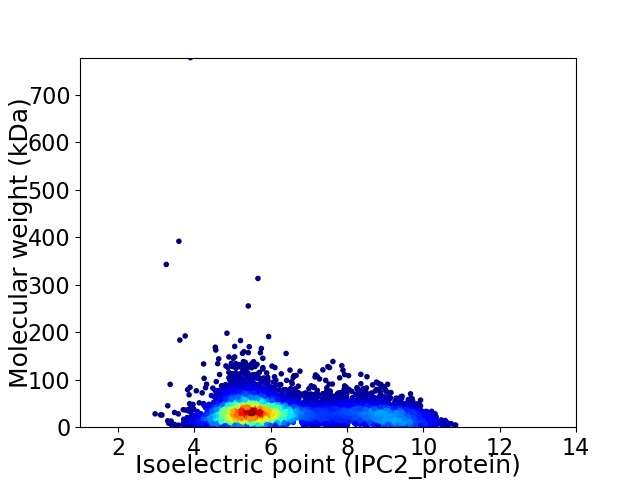
Virtual 2D-PAGE plot for 7269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W9H9D2|W9H9D2_9PROT Molybdopterin molybdenumtransferase OS=Skermanella stibiiresistens SB22 OX=1385369 GN=N825_34615 PE=3 SV=1
MM1 pKa = 7.63AGAALAVAHH10 pKa = 5.66SHH12 pKa = 6.46GALAQMTVKK21 pKa = 10.77LGGYY25 pKa = 4.57TTFRR29 pKa = 11.84AATYY33 pKa = 10.8DD34 pKa = 3.2SDD36 pKa = 4.94APGATSRR43 pKa = 11.84EE44 pKa = 3.99FQNEE48 pKa = 4.05VEE50 pKa = 4.31IHH52 pKa = 5.47VSADD56 pKa = 2.95GKK58 pKa = 11.02ADD60 pKa = 3.39NGLLYY65 pKa = 10.34GAHH68 pKa = 6.89IEE70 pKa = 4.13FEE72 pKa = 4.27NDD74 pKa = 3.06GGDD77 pKa = 3.48GAGAGISVDD86 pKa = 3.51EE87 pKa = 4.43ASVYY91 pKa = 10.95VGGTWGRR98 pKa = 11.84IEE100 pKa = 5.8LGDD103 pKa = 3.89HH104 pKa = 7.37DD105 pKa = 4.71GASDD109 pKa = 3.53QLFVYY114 pKa = 10.49APTVGNGQIEE124 pKa = 4.38EE125 pKa = 4.16DD126 pKa = 3.71WYY128 pKa = 10.88DD129 pKa = 3.39FAGPGIEE136 pKa = 4.51TDD138 pKa = 4.78DD139 pKa = 4.69LLLPTDD145 pKa = 4.08TSDD148 pKa = 3.49STKK151 pKa = 8.53ITYY154 pKa = 7.51MSPVLGGFQAGVSYY168 pKa = 9.86TPQFDD173 pKa = 3.66SQGQDD178 pKa = 2.99VVRR181 pKa = 11.84FKK183 pKa = 10.12PTSPDD188 pKa = 3.11AEE190 pKa = 4.19NLYY193 pKa = 10.78KK194 pKa = 10.68DD195 pKa = 4.8LIEE198 pKa = 5.49AGAGYY203 pKa = 10.2KK204 pKa = 9.73RR205 pKa = 11.84DD206 pKa = 3.59IGPVSLVLGVGYY218 pKa = 9.98VHH220 pKa = 7.47GDD222 pKa = 3.18ANQGTDD228 pKa = 5.04AEE230 pKa = 5.65DD231 pKa = 3.44FDD233 pKa = 4.3AWGAGAQIGYY243 pKa = 10.18EE244 pKa = 4.11GFTIGGAYY252 pKa = 9.8NDD254 pKa = 4.56NGDD257 pKa = 4.04SLIEE261 pKa = 4.04TDD263 pKa = 4.74EE264 pKa = 4.67PDD266 pKa = 3.34AKK268 pKa = 10.37SWNVGVSYY276 pKa = 10.46EE277 pKa = 4.03ADD279 pKa = 3.34DD280 pKa = 3.72WGVAAQYY287 pKa = 10.94QKK289 pKa = 11.21VDD291 pKa = 3.55LPGRR295 pKa = 11.84KK296 pKa = 8.82INIYY300 pKa = 10.33GLGAAYY306 pKa = 9.61EE307 pKa = 4.14PAPGLTIGPEE317 pKa = 4.17VIFFDD322 pKa = 5.75DD323 pKa = 4.97DD324 pKa = 3.94DD325 pKa = 5.99PEE327 pKa = 5.27VGDD330 pKa = 4.72GYY332 pKa = 11.23VALIAVNLEE341 pKa = 4.03FF342 pKa = 6.0
MM1 pKa = 7.63AGAALAVAHH10 pKa = 5.66SHH12 pKa = 6.46GALAQMTVKK21 pKa = 10.77LGGYY25 pKa = 4.57TTFRR29 pKa = 11.84AATYY33 pKa = 10.8DD34 pKa = 3.2SDD36 pKa = 4.94APGATSRR43 pKa = 11.84EE44 pKa = 3.99FQNEE48 pKa = 4.05VEE50 pKa = 4.31IHH52 pKa = 5.47VSADD56 pKa = 2.95GKK58 pKa = 11.02ADD60 pKa = 3.39NGLLYY65 pKa = 10.34GAHH68 pKa = 6.89IEE70 pKa = 4.13FEE72 pKa = 4.27NDD74 pKa = 3.06GGDD77 pKa = 3.48GAGAGISVDD86 pKa = 3.51EE87 pKa = 4.43ASVYY91 pKa = 10.95VGGTWGRR98 pKa = 11.84IEE100 pKa = 5.8LGDD103 pKa = 3.89HH104 pKa = 7.37DD105 pKa = 4.71GASDD109 pKa = 3.53QLFVYY114 pKa = 10.49APTVGNGQIEE124 pKa = 4.38EE125 pKa = 4.16DD126 pKa = 3.71WYY128 pKa = 10.88DD129 pKa = 3.39FAGPGIEE136 pKa = 4.51TDD138 pKa = 4.78DD139 pKa = 4.69LLLPTDD145 pKa = 4.08TSDD148 pKa = 3.49STKK151 pKa = 8.53ITYY154 pKa = 7.51MSPVLGGFQAGVSYY168 pKa = 9.86TPQFDD173 pKa = 3.66SQGQDD178 pKa = 2.99VVRR181 pKa = 11.84FKK183 pKa = 10.12PTSPDD188 pKa = 3.11AEE190 pKa = 4.19NLYY193 pKa = 10.78KK194 pKa = 10.68DD195 pKa = 4.8LIEE198 pKa = 5.49AGAGYY203 pKa = 10.2KK204 pKa = 9.73RR205 pKa = 11.84DD206 pKa = 3.59IGPVSLVLGVGYY218 pKa = 9.98VHH220 pKa = 7.47GDD222 pKa = 3.18ANQGTDD228 pKa = 5.04AEE230 pKa = 5.65DD231 pKa = 3.44FDD233 pKa = 4.3AWGAGAQIGYY243 pKa = 10.18EE244 pKa = 4.11GFTIGGAYY252 pKa = 9.8NDD254 pKa = 4.56NGDD257 pKa = 4.04SLIEE261 pKa = 4.04TDD263 pKa = 4.74EE264 pKa = 4.67PDD266 pKa = 3.34AKK268 pKa = 10.37SWNVGVSYY276 pKa = 10.46EE277 pKa = 4.03ADD279 pKa = 3.34DD280 pKa = 3.72WGVAAQYY287 pKa = 10.94QKK289 pKa = 11.21VDD291 pKa = 3.55LPGRR295 pKa = 11.84KK296 pKa = 8.82INIYY300 pKa = 10.33GLGAAYY306 pKa = 9.61EE307 pKa = 4.14PAPGLTIGPEE317 pKa = 4.17VIFFDD322 pKa = 5.75DD323 pKa = 4.97DD324 pKa = 3.94DD325 pKa = 5.99PEE327 pKa = 5.27VGDD330 pKa = 4.72GYY332 pKa = 11.23VALIAVNLEE341 pKa = 4.03FF342 pKa = 6.0
Molecular weight: 35.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W9GX22|W9GX22_9PROT Uncharacterized protein OS=Skermanella stibiiresistens SB22 OX=1385369 GN=N825_21450 PE=4 SV=1
MM1 pKa = 7.57ASSAAVRR8 pKa = 11.84FNILAAQIRR17 pKa = 11.84MMLTFFRR24 pKa = 11.84VSRR27 pKa = 11.84FFVLRR32 pKa = 3.85
MM1 pKa = 7.57ASSAAVRR8 pKa = 11.84FNILAAQIRR17 pKa = 11.84MMLTFFRR24 pKa = 11.84VSRR27 pKa = 11.84FFVLRR32 pKa = 3.85
Molecular weight: 3.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2267981 |
26 |
8112 |
312.0 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.259 ± 0.041 | 0.824 ± 0.011 |
5.94 ± 0.027 | 5.538 ± 0.029 |
3.512 ± 0.02 | 8.953 ± 0.032 |
2.09 ± 0.016 | 5.072 ± 0.021 |
2.813 ± 0.023 | 10.344 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.534 ± 0.016 | 2.44 ± 0.018 |
5.368 ± 0.026 | 2.896 ± 0.018 |
7.462 ± 0.04 | 5.226 ± 0.027 |
5.597 ± 0.048 | 7.812 ± 0.023 |
1.3 ± 0.012 | 2.019 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
