
Apis mellifera associated microvirus 60
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
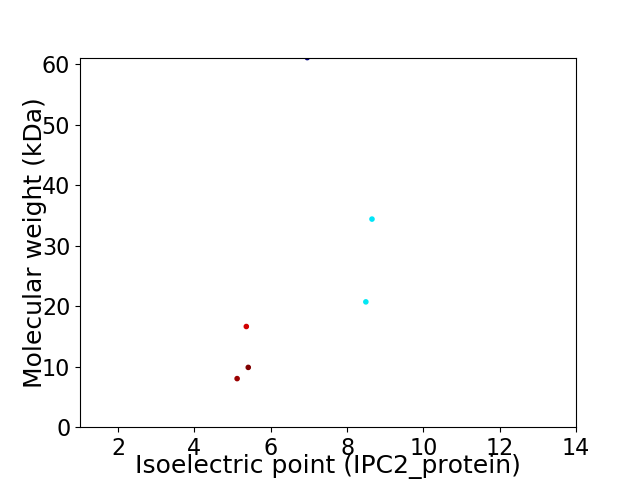
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
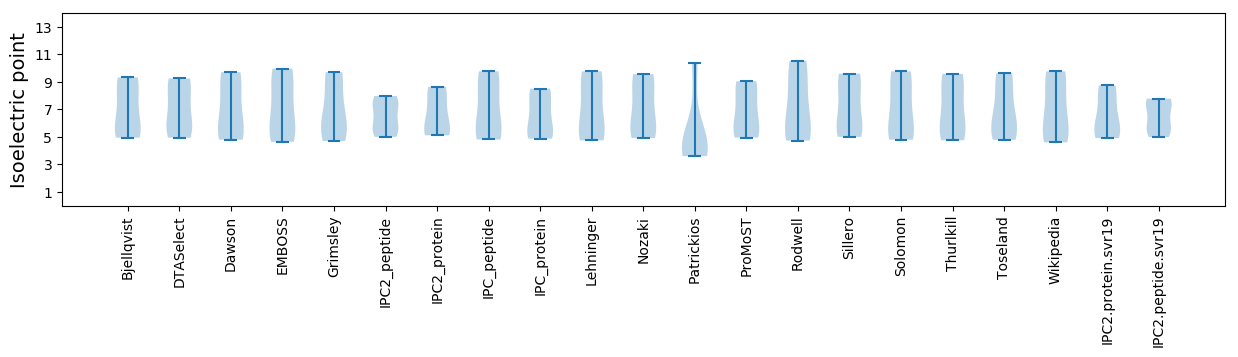
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U5E5|A0A3Q8U5E5_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 60 OX=2494792 PE=3 SV=1
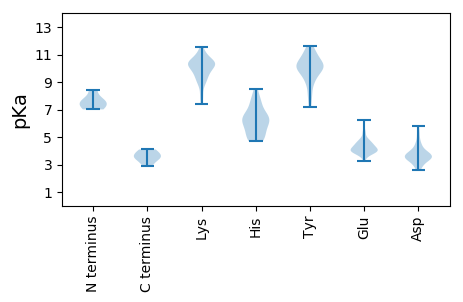
MM1 pKa = 8.46LEE3 pKa = 3.89EE4 pKa = 3.93QGILEE9 pKa = 4.37IVLQKK14 pKa = 10.7NCNITTTPKK23 pKa = 9.6QHH25 pKa = 6.28PTGGAASATEE35 pKa = 4.18AEE37 pKa = 4.45SPEE40 pKa = 3.89YY41 pKa = 10.39LYY43 pKa = 11.42YY44 pKa = 11.2LLIIFRR50 pKa = 11.84TDD52 pKa = 2.57TFYY55 pKa = 10.56LTKK58 pKa = 10.26SEE60 pKa = 4.26KK61 pKa = 9.8VSKK64 pKa = 10.57NIQQEE69 pKa = 4.59VVV71 pKa = 2.93
MM1 pKa = 8.46LEE3 pKa = 3.89EE4 pKa = 3.93QGILEE9 pKa = 4.37IVLQKK14 pKa = 10.7NCNITTTPKK23 pKa = 9.6QHH25 pKa = 6.28PTGGAASATEE35 pKa = 4.18AEE37 pKa = 4.45SPEE40 pKa = 3.89YY41 pKa = 10.39LYY43 pKa = 11.42YY44 pKa = 11.2LLIIFRR50 pKa = 11.84TDD52 pKa = 2.57TFYY55 pKa = 10.56LTKK58 pKa = 10.26SEE60 pKa = 4.26KK61 pKa = 9.8VSKK64 pKa = 10.57NIQQEE69 pKa = 4.59VVV71 pKa = 2.93
Molecular weight: 8.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTG4|A0A3S8UTG4_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 60 OX=2494792 PE=4 SV=1
MM1 pKa = 7.01CTKK4 pKa = 10.11PLKK7 pKa = 10.39AYY9 pKa = 9.32QNPEE13 pKa = 3.25GGRR16 pKa = 11.84PIFGYY21 pKa = 9.99QGVRR25 pKa = 11.84EE26 pKa = 4.73GLPEE30 pKa = 3.85MQLPCGKK37 pKa = 10.23CSEE40 pKa = 4.48CVKK43 pKa = 10.56DD44 pKa = 5.79YY45 pKa = 9.08YY46 pKa = 9.6TAWATRR52 pKa = 11.84GFRR55 pKa = 11.84EE56 pKa = 4.06LVQWEE61 pKa = 4.04NSVFITLTYY70 pKa = 11.01DD71 pKa = 3.42NEE73 pKa = 4.26NLPGDD78 pKa = 3.81YY79 pKa = 10.37SLKK82 pKa = 10.81KK83 pKa = 10.17KK84 pKa = 10.38DD85 pKa = 3.36VQDD88 pKa = 4.41FIKK91 pKa = 10.34RR92 pKa = 11.84VKK94 pKa = 10.1KK95 pKa = 9.82HH96 pKa = 5.23FRR98 pKa = 11.84SHH100 pKa = 6.46KK101 pKa = 8.84EE102 pKa = 3.55NPVRR106 pKa = 11.84QIYY109 pKa = 9.91CGEE112 pKa = 4.09YY113 pKa = 10.05GKK115 pKa = 8.77KK116 pKa = 7.61TLRR119 pKa = 11.84PHH121 pKa = 4.81YY122 pKa = 10.12HH123 pKa = 6.15VILFNVDD130 pKa = 3.71FKK132 pKa = 11.52DD133 pKa = 3.54KK134 pKa = 10.22KK135 pKa = 9.06KK136 pKa = 10.31HH137 pKa = 4.93YY138 pKa = 9.19MSDD141 pKa = 2.75GGKK144 pKa = 9.51QVFTSEE150 pKa = 3.71ILSRR154 pKa = 11.84LWGKK158 pKa = 9.2GNCDD162 pKa = 3.14FGYY165 pKa = 10.96AEE167 pKa = 4.92PEE169 pKa = 4.04SIAYY173 pKa = 8.92LFKK176 pKa = 11.15YY177 pKa = 9.51VLKK180 pKa = 10.7KK181 pKa = 9.27KK182 pKa = 7.6TRR184 pKa = 11.84KK185 pKa = 8.95EE186 pKa = 3.87RR187 pKa = 11.84EE188 pKa = 3.56KK189 pKa = 10.72PLYY192 pKa = 9.79HH193 pKa = 6.54EE194 pKa = 5.33HH195 pKa = 7.48DD196 pKa = 3.56GCIYY200 pKa = 9.55EE201 pKa = 4.73CEE203 pKa = 4.26HH204 pKa = 6.36EE205 pKa = 4.86FIEE208 pKa = 4.92ASRR211 pKa = 11.84NPGIGACAKK220 pKa = 10.09DD221 pKa = 3.58SPSIRR226 pKa = 11.84KK227 pKa = 9.07GFLVTNGVKK236 pKa = 10.33SKK238 pKa = 10.67LPKK241 pKa = 10.4YY242 pKa = 10.25YY243 pKa = 10.56LEE245 pKa = 4.49HH246 pKa = 6.68LKK248 pKa = 9.07KK249 pKa = 9.18TDD251 pKa = 3.38LPKK254 pKa = 10.57FLEE257 pKa = 4.5IQDD260 pKa = 3.71KK261 pKa = 10.48RR262 pKa = 11.84ATFALEE268 pKa = 3.93KK269 pKa = 10.28TPEE272 pKa = 3.83TGKK275 pKa = 10.26RR276 pKa = 11.84RR277 pKa = 11.84EE278 pKa = 4.13QKK280 pKa = 10.72EE281 pKa = 4.01EE282 pKa = 3.84ALKK285 pKa = 11.15LLTDD289 pKa = 3.99TKK291 pKa = 11.19RR292 pKa = 11.84RR293 pKa = 11.84MM294 pKa = 3.39
MM1 pKa = 7.01CTKK4 pKa = 10.11PLKK7 pKa = 10.39AYY9 pKa = 9.32QNPEE13 pKa = 3.25GGRR16 pKa = 11.84PIFGYY21 pKa = 9.99QGVRR25 pKa = 11.84EE26 pKa = 4.73GLPEE30 pKa = 3.85MQLPCGKK37 pKa = 10.23CSEE40 pKa = 4.48CVKK43 pKa = 10.56DD44 pKa = 5.79YY45 pKa = 9.08YY46 pKa = 9.6TAWATRR52 pKa = 11.84GFRR55 pKa = 11.84EE56 pKa = 4.06LVQWEE61 pKa = 4.04NSVFITLTYY70 pKa = 11.01DD71 pKa = 3.42NEE73 pKa = 4.26NLPGDD78 pKa = 3.81YY79 pKa = 10.37SLKK82 pKa = 10.81KK83 pKa = 10.17KK84 pKa = 10.38DD85 pKa = 3.36VQDD88 pKa = 4.41FIKK91 pKa = 10.34RR92 pKa = 11.84VKK94 pKa = 10.1KK95 pKa = 9.82HH96 pKa = 5.23FRR98 pKa = 11.84SHH100 pKa = 6.46KK101 pKa = 8.84EE102 pKa = 3.55NPVRR106 pKa = 11.84QIYY109 pKa = 9.91CGEE112 pKa = 4.09YY113 pKa = 10.05GKK115 pKa = 8.77KK116 pKa = 7.61TLRR119 pKa = 11.84PHH121 pKa = 4.81YY122 pKa = 10.12HH123 pKa = 6.15VILFNVDD130 pKa = 3.71FKK132 pKa = 11.52DD133 pKa = 3.54KK134 pKa = 10.22KK135 pKa = 9.06KK136 pKa = 10.31HH137 pKa = 4.93YY138 pKa = 9.19MSDD141 pKa = 2.75GGKK144 pKa = 9.51QVFTSEE150 pKa = 3.71ILSRR154 pKa = 11.84LWGKK158 pKa = 9.2GNCDD162 pKa = 3.14FGYY165 pKa = 10.96AEE167 pKa = 4.92PEE169 pKa = 4.04SIAYY173 pKa = 8.92LFKK176 pKa = 11.15YY177 pKa = 9.51VLKK180 pKa = 10.7KK181 pKa = 9.27KK182 pKa = 7.6TRR184 pKa = 11.84KK185 pKa = 8.95EE186 pKa = 3.87RR187 pKa = 11.84EE188 pKa = 3.56KK189 pKa = 10.72PLYY192 pKa = 9.79HH193 pKa = 6.54EE194 pKa = 5.33HH195 pKa = 7.48DD196 pKa = 3.56GCIYY200 pKa = 9.55EE201 pKa = 4.73CEE203 pKa = 4.26HH204 pKa = 6.36EE205 pKa = 4.86FIEE208 pKa = 4.92ASRR211 pKa = 11.84NPGIGACAKK220 pKa = 10.09DD221 pKa = 3.58SPSIRR226 pKa = 11.84KK227 pKa = 9.07GFLVTNGVKK236 pKa = 10.33SKK238 pKa = 10.67LPKK241 pKa = 10.4YY242 pKa = 10.25YY243 pKa = 10.56LEE245 pKa = 4.49HH246 pKa = 6.68LKK248 pKa = 9.07KK249 pKa = 9.18TDD251 pKa = 3.38LPKK254 pKa = 10.57FLEE257 pKa = 4.5IQDD260 pKa = 3.71KK261 pKa = 10.48RR262 pKa = 11.84ATFALEE268 pKa = 3.93KK269 pKa = 10.28TPEE272 pKa = 3.83TGKK275 pKa = 10.26RR276 pKa = 11.84RR277 pKa = 11.84EE278 pKa = 4.13QKK280 pKa = 10.72EE281 pKa = 4.01EE282 pKa = 3.84ALKK285 pKa = 11.15LLTDD289 pKa = 3.99TKK291 pKa = 11.19RR292 pKa = 11.84RR293 pKa = 11.84MM294 pKa = 3.39
Molecular weight: 34.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1345 |
71 |
552 |
224.2 |
25.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.361 ± 1.511 | 1.19 ± 0.523 |
5.13 ± 0.566 | 6.691 ± 1.542 |
3.569 ± 0.804 | 7.138 ± 0.663 |
1.859 ± 0.537 | 5.502 ± 0.706 |
7.435 ± 2.343 | 8.476 ± 0.456 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.528 ± 0.412 | 4.312 ± 0.664 |
5.056 ± 0.394 | 5.651 ± 0.889 |
5.13 ± 0.999 | 6.32 ± 0.85 |
6.989 ± 0.977 | 4.833 ± 0.486 |
0.892 ± 0.217 | 3.941 ± 0.76 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
