
Beihai permutotetra-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
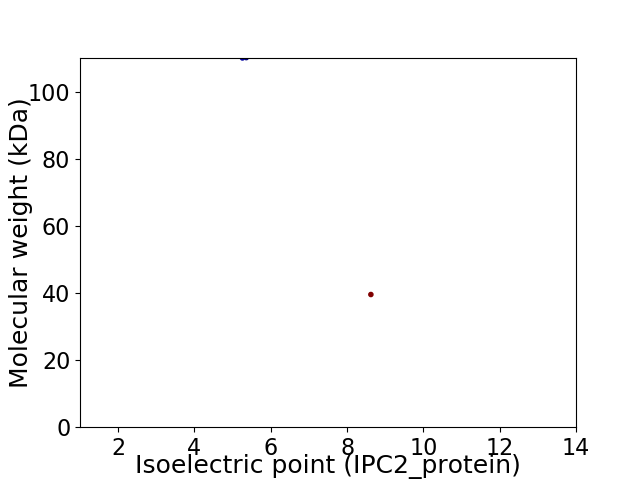
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHU9|A0A1L3KHU9_9VIRU RdRp OS=Beihai permutotetra-like virus 2 OX=1922513 PE=4 SV=1
MM1 pKa = 7.87DD2 pKa = 5.44ASNPVSSGDD11 pKa = 3.35KK12 pKa = 9.72VTFADD17 pKa = 4.27LKK19 pKa = 10.86EE20 pKa = 4.61DD21 pKa = 3.5SLLLKK26 pKa = 10.12RR27 pKa = 11.84QNFEE31 pKa = 3.98EE32 pKa = 4.18LKK34 pKa = 10.65KK35 pKa = 11.14LIGTSPRR42 pKa = 11.84TPAPYY47 pKa = 10.01VGRR50 pKa = 11.84NEE52 pKa = 3.67YY53 pKa = 10.68DD54 pKa = 3.3YY55 pKa = 11.83DD56 pKa = 4.17EE57 pKa = 6.01DD58 pKa = 4.58VVKK61 pKa = 10.99DD62 pKa = 4.35LIKK65 pKa = 10.56PIAVAKK71 pKa = 9.9GNPEE75 pKa = 3.84VGFLTEE81 pKa = 3.7AAKK84 pKa = 10.77VRR86 pKa = 11.84VCSFHH91 pKa = 6.77TMGEE95 pKa = 4.1NQEE98 pKa = 4.37DD99 pKa = 4.15NNVSMKK105 pKa = 8.78QTYY108 pKa = 10.0KK109 pKa = 9.54IAKK112 pKa = 8.67RR113 pKa = 11.84DD114 pKa = 3.5RR115 pKa = 11.84KK116 pKa = 10.23LVEE119 pKa = 3.88VCLKK123 pKa = 10.82SGAQSDD129 pKa = 4.1DD130 pKa = 3.1IKK132 pKa = 11.16RR133 pKa = 11.84LIYY136 pKa = 8.2TTGNKK141 pKa = 9.8EE142 pKa = 4.09GFSKK146 pKa = 10.68RR147 pKa = 11.84LRR149 pKa = 11.84LWATKK154 pKa = 9.84KK155 pKa = 10.6VPSPKK160 pKa = 9.64TAMEE164 pKa = 4.21KK165 pKa = 10.71YY166 pKa = 9.52NIKK169 pKa = 10.67FEE171 pKa = 4.41DD172 pKa = 3.35VRR174 pKa = 11.84AALPVGKK181 pKa = 10.14GLPNWNGNIEE191 pKa = 4.08EE192 pKa = 4.81FISGVRR198 pKa = 11.84VNKK201 pKa = 10.45SSGAGPPFYY210 pKa = 9.65RR211 pKa = 11.84TKK213 pKa = 10.51RR214 pKa = 11.84QCLDD218 pKa = 3.35DD219 pKa = 4.76CFDD222 pKa = 6.11AIGKK226 pKa = 8.84LIKK229 pKa = 9.9EE230 pKa = 4.48ANQDD234 pKa = 3.59HH235 pKa = 7.2LDD237 pKa = 3.31KK238 pKa = 10.78FLRR241 pKa = 11.84EE242 pKa = 3.95NEE244 pKa = 4.1EE245 pKa = 4.72FLVCQCKK252 pKa = 10.52NKK254 pKa = 8.99TDD256 pKa = 3.94RR257 pKa = 11.84YY258 pKa = 9.89PPDD261 pKa = 3.56KK262 pKa = 10.52LHH264 pKa = 6.79EE265 pKa = 4.07KK266 pKa = 8.13TRR268 pKa = 11.84PYY270 pKa = 10.79FSFSFPTQFLCSALNQPFTDD290 pKa = 3.38NMLLFHH296 pKa = 7.15EE297 pKa = 5.33SGANACGFSYY307 pKa = 11.4ADD309 pKa = 3.91GGSQRR314 pKa = 11.84LFDD317 pKa = 3.79WMCDD321 pKa = 3.46TEE323 pKa = 4.15EE324 pKa = 6.03GEE326 pKa = 5.22VKK328 pKa = 10.45FCAYY332 pKa = 10.38GDD334 pKa = 3.7DD335 pKa = 3.31VRR337 pKa = 11.84MVKK340 pKa = 10.38RR341 pKa = 11.84EE342 pKa = 3.94DD343 pKa = 3.44GVLYY347 pKa = 10.61SCSPDD352 pKa = 3.69FQCMDD357 pKa = 3.31GSIHH361 pKa = 5.45KK362 pKa = 8.91TEE364 pKa = 3.69AAEE367 pKa = 3.96YY368 pKa = 9.43VDD370 pKa = 5.28YY371 pKa = 11.24VLHH374 pKa = 6.52CYY376 pKa = 8.7EE377 pKa = 4.87RR378 pKa = 11.84EE379 pKa = 4.09HH380 pKa = 6.96GPSNFWKK387 pKa = 10.57FVGEE391 pKa = 4.09VWKK394 pKa = 10.36KK395 pKa = 9.59QLVGAKK401 pKa = 9.45FFVNGSQPYY410 pKa = 9.38RR411 pKa = 11.84SEE413 pKa = 3.98TGLLTGCVGTTGVDD427 pKa = 3.25TFKK430 pKa = 11.19SVTAFVTLIEE440 pKa = 4.09AHH442 pKa = 5.91KK443 pKa = 10.16HH444 pKa = 4.64YY445 pKa = 10.69GVDD448 pKa = 2.9IMNVEE453 pKa = 4.08EE454 pKa = 4.57VKK456 pKa = 11.21NFMRR460 pKa = 11.84EE461 pKa = 3.76NFGLVLKK468 pKa = 10.71GDD470 pKa = 4.2TYY472 pKa = 11.39QWDD475 pKa = 3.91EE476 pKa = 4.04VQEE479 pKa = 3.97EE480 pKa = 4.69LEE482 pKa = 4.5EE483 pKa = 4.82GEE485 pKa = 4.3QPNAQEE491 pKa = 3.99FLGVQLKK498 pKa = 10.57AVTGKK503 pKa = 9.82NRR505 pKa = 11.84LEE507 pKa = 4.05VIPYY511 pKa = 9.66KK512 pKa = 10.54EE513 pKa = 3.96EE514 pKa = 4.01SDD516 pKa = 3.61IVSLIANARR525 pKa = 11.84VPEE528 pKa = 4.07EE529 pKa = 3.78VRR531 pKa = 11.84DD532 pKa = 3.88LKK534 pKa = 10.62KK535 pKa = 7.39TTGRR539 pKa = 11.84ARR541 pKa = 11.84YY542 pKa = 9.49LFDD545 pKa = 3.59CARR548 pKa = 11.84GYY550 pKa = 10.25MITAAFLHH558 pKa = 6.2EE559 pKa = 4.58KK560 pKa = 8.01TWNICSRR567 pKa = 11.84LIEE570 pKa = 4.13EE571 pKa = 4.7TNTEE575 pKa = 4.52VICMRR580 pKa = 11.84VQTGKK585 pKa = 10.76KK586 pKa = 9.65VDD588 pKa = 3.52GEE590 pKa = 4.44YY591 pKa = 10.9GGEE594 pKa = 4.19GPEE597 pKa = 4.2LVDD600 pKa = 5.52LVGEE604 pKa = 4.3DD605 pKa = 4.17FTWPSSDD612 pKa = 2.77GWPSIDD618 pKa = 3.59FCVDD622 pKa = 3.18VYY624 pKa = 11.06LSPDD628 pKa = 3.29NKK630 pKa = 10.48RR631 pKa = 11.84GGLWYY636 pKa = 10.19DD637 pKa = 3.4CVPALRR643 pKa = 11.84EE644 pKa = 3.82KK645 pKa = 10.77LDD647 pKa = 3.84EE648 pKa = 4.22IKK650 pKa = 10.35RR651 pKa = 11.84YY652 pKa = 9.66RR653 pKa = 11.84SLRR656 pKa = 11.84DD657 pKa = 3.3PHH659 pKa = 6.79HH660 pKa = 6.53VPPVVSRR667 pKa = 11.84EE668 pKa = 3.98VQTTSWAEE676 pKa = 3.74EE677 pKa = 4.27VEE679 pKa = 4.4TEE681 pKa = 4.22RR682 pKa = 11.84GRR684 pKa = 11.84EE685 pKa = 3.83QAEE688 pKa = 3.71LAMEE692 pKa = 4.93RR693 pKa = 11.84KK694 pKa = 9.54RR695 pKa = 11.84DD696 pKa = 3.73PTVDD700 pKa = 3.06SEE702 pKa = 4.95GYY704 pKa = 9.73LRR706 pKa = 11.84SFLEE710 pKa = 3.87EE711 pKa = 3.88RR712 pKa = 11.84TPLMKK717 pKa = 9.9PVKK720 pKa = 9.84YY721 pKa = 9.56PKK723 pKa = 10.52EE724 pKa = 3.84FVKK727 pKa = 11.04YY728 pKa = 10.0KK729 pKa = 10.04ATTQSVPKK737 pKa = 9.81EE738 pKa = 3.87KK739 pKa = 9.95RR740 pKa = 11.84IEE742 pKa = 3.89NAIPEE747 pKa = 4.5GAEE750 pKa = 3.69KK751 pKa = 10.18FHH753 pKa = 7.39RR754 pKa = 11.84DD755 pKa = 4.72FIPLVTNLAPGKK767 pKa = 9.32CAEE770 pKa = 4.2VLISLGWRR778 pKa = 11.84PSPSEE783 pKa = 4.34SYY785 pKa = 9.12WEE787 pKa = 4.26KK788 pKa = 11.16GPLVPVRR795 pKa = 11.84DD796 pKa = 4.15SDD798 pKa = 3.62WTRR801 pKa = 11.84AEE803 pKa = 4.1LDD805 pKa = 3.33RR806 pKa = 11.84LKK808 pKa = 10.49EE809 pKa = 3.97VPRR812 pKa = 11.84RR813 pKa = 11.84EE814 pKa = 3.92TKK816 pKa = 10.71SRR818 pKa = 11.84VKK820 pKa = 10.26VARR823 pKa = 11.84DD824 pKa = 3.04GKK826 pKa = 7.34TWCEE830 pKa = 3.85VPDD833 pKa = 4.06DD834 pKa = 4.24VKK836 pKa = 11.35LSDD839 pKa = 3.57RR840 pKa = 11.84QVDD843 pKa = 3.94VVSRR847 pKa = 11.84VSYY850 pKa = 10.21SFVNAGHH857 pKa = 6.59ILDD860 pKa = 3.76RR861 pKa = 11.84KK862 pKa = 8.64YY863 pKa = 9.28TVLEE867 pKa = 3.98NSPNPWGVLQVTYY880 pKa = 10.56RR881 pKa = 11.84GTGVLVGEE889 pKa = 4.52GFGVKK894 pKa = 9.48KK895 pKa = 10.81DD896 pKa = 3.65EE897 pKa = 4.18ATSAVFEE904 pKa = 4.55LMMEE908 pKa = 4.68RR909 pKa = 11.84IMAKK913 pKa = 9.84EE914 pKa = 3.89AGPLTSEE921 pKa = 4.66IVTTEE926 pKa = 3.61NGEE929 pKa = 4.0NRR931 pKa = 11.84ATEE934 pKa = 4.06SEE936 pKa = 4.11AARR939 pKa = 11.84EE940 pKa = 4.02EE941 pKa = 4.28EE942 pKa = 4.05TAEE945 pKa = 4.13TEE947 pKa = 4.44RR948 pKa = 11.84EE949 pKa = 3.98TEE951 pKa = 3.94EE952 pKa = 4.4RR953 pKa = 11.84NEE955 pKa = 4.22SGSTQTVAPASEE967 pKa = 4.38GKK969 pKa = 10.32DD970 pKa = 3.29GGNN973 pKa = 3.33
MM1 pKa = 7.87DD2 pKa = 5.44ASNPVSSGDD11 pKa = 3.35KK12 pKa = 9.72VTFADD17 pKa = 4.27LKK19 pKa = 10.86EE20 pKa = 4.61DD21 pKa = 3.5SLLLKK26 pKa = 10.12RR27 pKa = 11.84QNFEE31 pKa = 3.98EE32 pKa = 4.18LKK34 pKa = 10.65KK35 pKa = 11.14LIGTSPRR42 pKa = 11.84TPAPYY47 pKa = 10.01VGRR50 pKa = 11.84NEE52 pKa = 3.67YY53 pKa = 10.68DD54 pKa = 3.3YY55 pKa = 11.83DD56 pKa = 4.17EE57 pKa = 6.01DD58 pKa = 4.58VVKK61 pKa = 10.99DD62 pKa = 4.35LIKK65 pKa = 10.56PIAVAKK71 pKa = 9.9GNPEE75 pKa = 3.84VGFLTEE81 pKa = 3.7AAKK84 pKa = 10.77VRR86 pKa = 11.84VCSFHH91 pKa = 6.77TMGEE95 pKa = 4.1NQEE98 pKa = 4.37DD99 pKa = 4.15NNVSMKK105 pKa = 8.78QTYY108 pKa = 10.0KK109 pKa = 9.54IAKK112 pKa = 8.67RR113 pKa = 11.84DD114 pKa = 3.5RR115 pKa = 11.84KK116 pKa = 10.23LVEE119 pKa = 3.88VCLKK123 pKa = 10.82SGAQSDD129 pKa = 4.1DD130 pKa = 3.1IKK132 pKa = 11.16RR133 pKa = 11.84LIYY136 pKa = 8.2TTGNKK141 pKa = 9.8EE142 pKa = 4.09GFSKK146 pKa = 10.68RR147 pKa = 11.84LRR149 pKa = 11.84LWATKK154 pKa = 9.84KK155 pKa = 10.6VPSPKK160 pKa = 9.64TAMEE164 pKa = 4.21KK165 pKa = 10.71YY166 pKa = 9.52NIKK169 pKa = 10.67FEE171 pKa = 4.41DD172 pKa = 3.35VRR174 pKa = 11.84AALPVGKK181 pKa = 10.14GLPNWNGNIEE191 pKa = 4.08EE192 pKa = 4.81FISGVRR198 pKa = 11.84VNKK201 pKa = 10.45SSGAGPPFYY210 pKa = 9.65RR211 pKa = 11.84TKK213 pKa = 10.51RR214 pKa = 11.84QCLDD218 pKa = 3.35DD219 pKa = 4.76CFDD222 pKa = 6.11AIGKK226 pKa = 8.84LIKK229 pKa = 9.9EE230 pKa = 4.48ANQDD234 pKa = 3.59HH235 pKa = 7.2LDD237 pKa = 3.31KK238 pKa = 10.78FLRR241 pKa = 11.84EE242 pKa = 3.95NEE244 pKa = 4.1EE245 pKa = 4.72FLVCQCKK252 pKa = 10.52NKK254 pKa = 8.99TDD256 pKa = 3.94RR257 pKa = 11.84YY258 pKa = 9.89PPDD261 pKa = 3.56KK262 pKa = 10.52LHH264 pKa = 6.79EE265 pKa = 4.07KK266 pKa = 8.13TRR268 pKa = 11.84PYY270 pKa = 10.79FSFSFPTQFLCSALNQPFTDD290 pKa = 3.38NMLLFHH296 pKa = 7.15EE297 pKa = 5.33SGANACGFSYY307 pKa = 11.4ADD309 pKa = 3.91GGSQRR314 pKa = 11.84LFDD317 pKa = 3.79WMCDD321 pKa = 3.46TEE323 pKa = 4.15EE324 pKa = 6.03GEE326 pKa = 5.22VKK328 pKa = 10.45FCAYY332 pKa = 10.38GDD334 pKa = 3.7DD335 pKa = 3.31VRR337 pKa = 11.84MVKK340 pKa = 10.38RR341 pKa = 11.84EE342 pKa = 3.94DD343 pKa = 3.44GVLYY347 pKa = 10.61SCSPDD352 pKa = 3.69FQCMDD357 pKa = 3.31GSIHH361 pKa = 5.45KK362 pKa = 8.91TEE364 pKa = 3.69AAEE367 pKa = 3.96YY368 pKa = 9.43VDD370 pKa = 5.28YY371 pKa = 11.24VLHH374 pKa = 6.52CYY376 pKa = 8.7EE377 pKa = 4.87RR378 pKa = 11.84EE379 pKa = 4.09HH380 pKa = 6.96GPSNFWKK387 pKa = 10.57FVGEE391 pKa = 4.09VWKK394 pKa = 10.36KK395 pKa = 9.59QLVGAKK401 pKa = 9.45FFVNGSQPYY410 pKa = 9.38RR411 pKa = 11.84SEE413 pKa = 3.98TGLLTGCVGTTGVDD427 pKa = 3.25TFKK430 pKa = 11.19SVTAFVTLIEE440 pKa = 4.09AHH442 pKa = 5.91KK443 pKa = 10.16HH444 pKa = 4.64YY445 pKa = 10.69GVDD448 pKa = 2.9IMNVEE453 pKa = 4.08EE454 pKa = 4.57VKK456 pKa = 11.21NFMRR460 pKa = 11.84EE461 pKa = 3.76NFGLVLKK468 pKa = 10.71GDD470 pKa = 4.2TYY472 pKa = 11.39QWDD475 pKa = 3.91EE476 pKa = 4.04VQEE479 pKa = 3.97EE480 pKa = 4.69LEE482 pKa = 4.5EE483 pKa = 4.82GEE485 pKa = 4.3QPNAQEE491 pKa = 3.99FLGVQLKK498 pKa = 10.57AVTGKK503 pKa = 9.82NRR505 pKa = 11.84LEE507 pKa = 4.05VIPYY511 pKa = 9.66KK512 pKa = 10.54EE513 pKa = 3.96EE514 pKa = 4.01SDD516 pKa = 3.61IVSLIANARR525 pKa = 11.84VPEE528 pKa = 4.07EE529 pKa = 3.78VRR531 pKa = 11.84DD532 pKa = 3.88LKK534 pKa = 10.62KK535 pKa = 7.39TTGRR539 pKa = 11.84ARR541 pKa = 11.84YY542 pKa = 9.49LFDD545 pKa = 3.59CARR548 pKa = 11.84GYY550 pKa = 10.25MITAAFLHH558 pKa = 6.2EE559 pKa = 4.58KK560 pKa = 8.01TWNICSRR567 pKa = 11.84LIEE570 pKa = 4.13EE571 pKa = 4.7TNTEE575 pKa = 4.52VICMRR580 pKa = 11.84VQTGKK585 pKa = 10.76KK586 pKa = 9.65VDD588 pKa = 3.52GEE590 pKa = 4.44YY591 pKa = 10.9GGEE594 pKa = 4.19GPEE597 pKa = 4.2LVDD600 pKa = 5.52LVGEE604 pKa = 4.3DD605 pKa = 4.17FTWPSSDD612 pKa = 2.77GWPSIDD618 pKa = 3.59FCVDD622 pKa = 3.18VYY624 pKa = 11.06LSPDD628 pKa = 3.29NKK630 pKa = 10.48RR631 pKa = 11.84GGLWYY636 pKa = 10.19DD637 pKa = 3.4CVPALRR643 pKa = 11.84EE644 pKa = 3.82KK645 pKa = 10.77LDD647 pKa = 3.84EE648 pKa = 4.22IKK650 pKa = 10.35RR651 pKa = 11.84YY652 pKa = 9.66RR653 pKa = 11.84SLRR656 pKa = 11.84DD657 pKa = 3.3PHH659 pKa = 6.79HH660 pKa = 6.53VPPVVSRR667 pKa = 11.84EE668 pKa = 3.98VQTTSWAEE676 pKa = 3.74EE677 pKa = 4.27VEE679 pKa = 4.4TEE681 pKa = 4.22RR682 pKa = 11.84GRR684 pKa = 11.84EE685 pKa = 3.83QAEE688 pKa = 3.71LAMEE692 pKa = 4.93RR693 pKa = 11.84KK694 pKa = 9.54RR695 pKa = 11.84DD696 pKa = 3.73PTVDD700 pKa = 3.06SEE702 pKa = 4.95GYY704 pKa = 9.73LRR706 pKa = 11.84SFLEE710 pKa = 3.87EE711 pKa = 3.88RR712 pKa = 11.84TPLMKK717 pKa = 9.9PVKK720 pKa = 9.84YY721 pKa = 9.56PKK723 pKa = 10.52EE724 pKa = 3.84FVKK727 pKa = 11.04YY728 pKa = 10.0KK729 pKa = 10.04ATTQSVPKK737 pKa = 9.81EE738 pKa = 3.87KK739 pKa = 9.95RR740 pKa = 11.84IEE742 pKa = 3.89NAIPEE747 pKa = 4.5GAEE750 pKa = 3.69KK751 pKa = 10.18FHH753 pKa = 7.39RR754 pKa = 11.84DD755 pKa = 4.72FIPLVTNLAPGKK767 pKa = 9.32CAEE770 pKa = 4.2VLISLGWRR778 pKa = 11.84PSPSEE783 pKa = 4.34SYY785 pKa = 9.12WEE787 pKa = 4.26KK788 pKa = 11.16GPLVPVRR795 pKa = 11.84DD796 pKa = 4.15SDD798 pKa = 3.62WTRR801 pKa = 11.84AEE803 pKa = 4.1LDD805 pKa = 3.33RR806 pKa = 11.84LKK808 pKa = 10.49EE809 pKa = 3.97VPRR812 pKa = 11.84RR813 pKa = 11.84EE814 pKa = 3.92TKK816 pKa = 10.71SRR818 pKa = 11.84VKK820 pKa = 10.26VARR823 pKa = 11.84DD824 pKa = 3.04GKK826 pKa = 7.34TWCEE830 pKa = 3.85VPDD833 pKa = 4.06DD834 pKa = 4.24VKK836 pKa = 11.35LSDD839 pKa = 3.57RR840 pKa = 11.84QVDD843 pKa = 3.94VVSRR847 pKa = 11.84VSYY850 pKa = 10.21SFVNAGHH857 pKa = 6.59ILDD860 pKa = 3.76RR861 pKa = 11.84KK862 pKa = 8.64YY863 pKa = 9.28TVLEE867 pKa = 3.98NSPNPWGVLQVTYY880 pKa = 10.56RR881 pKa = 11.84GTGVLVGEE889 pKa = 4.52GFGVKK894 pKa = 9.48KK895 pKa = 10.81DD896 pKa = 3.65EE897 pKa = 4.18ATSAVFEE904 pKa = 4.55LMMEE908 pKa = 4.68RR909 pKa = 11.84IMAKK913 pKa = 9.84EE914 pKa = 3.89AGPLTSEE921 pKa = 4.66IVTTEE926 pKa = 3.61NGEE929 pKa = 4.0NRR931 pKa = 11.84ATEE934 pKa = 4.06SEE936 pKa = 4.11AARR939 pKa = 11.84EE940 pKa = 4.02EE941 pKa = 4.28EE942 pKa = 4.05TAEE945 pKa = 4.13TEE947 pKa = 4.44RR948 pKa = 11.84EE949 pKa = 3.98TEE951 pKa = 3.94EE952 pKa = 4.4RR953 pKa = 11.84NEE955 pKa = 4.22SGSTQTVAPASEE967 pKa = 4.38GKK969 pKa = 10.32DD970 pKa = 3.29GGNN973 pKa = 3.33
Molecular weight: 110.25 kDa
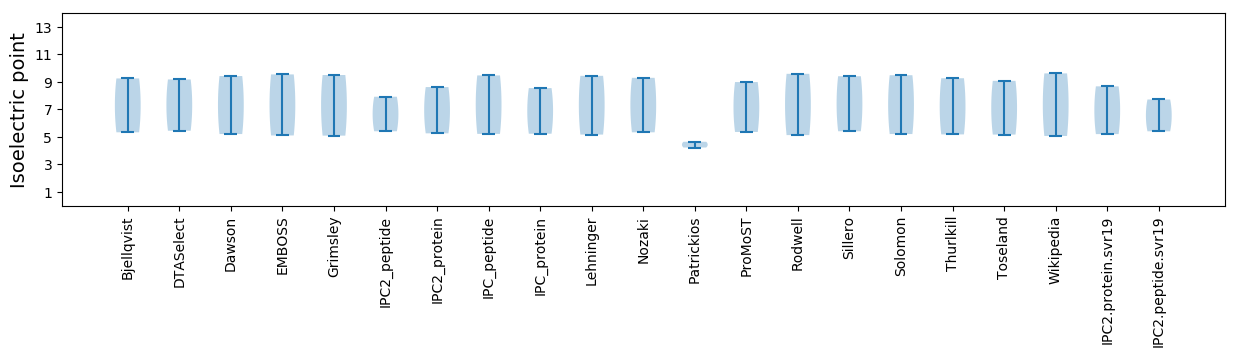
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHU9|A0A1L3KHU9_9VIRU RdRp OS=Beihai permutotetra-like virus 2 OX=1922513 PE=4 SV=1
MM1 pKa = 7.41VKK3 pKa = 9.41TEE5 pKa = 3.87QQRR8 pKa = 11.84AKK10 pKa = 9.79QRR12 pKa = 11.84EE13 pKa = 3.62RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84QRR19 pKa = 11.84QNARR23 pKa = 11.84LRR25 pKa = 11.84KK26 pKa = 9.34EE27 pKa = 4.21MNQEE31 pKa = 3.73APRR34 pKa = 11.84PLPQRR39 pKa = 11.84QRR41 pKa = 11.84GRR43 pKa = 11.84MGGIKK48 pKa = 10.07RR49 pKa = 11.84NCFDD53 pKa = 4.08GNDD56 pKa = 4.42LISSVDD62 pKa = 3.49VPATAKK68 pKa = 10.37SGTVIARR75 pKa = 11.84VDD77 pKa = 3.19ICPSKK82 pKa = 10.96FPGTSLWRR90 pKa = 11.84QAQLYY95 pKa = 9.8ANWKK99 pKa = 7.63PARR102 pKa = 11.84KK103 pKa = 9.27LKK105 pKa = 10.58IVVQPSAGTTTSGSYY120 pKa = 10.48IMGWTMDD127 pKa = 3.73SKK129 pKa = 11.7LDD131 pKa = 3.78IPSGDD136 pKa = 3.29NAVRR140 pKa = 11.84TVATFEE146 pKa = 3.8RR147 pKa = 11.84RR148 pKa = 11.84QTAKK152 pKa = 10.1IYY154 pKa = 9.41EE155 pKa = 4.32KK156 pKa = 10.93KK157 pKa = 10.31EE158 pKa = 3.6LNIPLKK164 pKa = 9.79TVQALLFTDD173 pKa = 3.93SSKK176 pKa = 11.17EE177 pKa = 3.93DD178 pKa = 3.37SDD180 pKa = 3.45QGSFYY185 pKa = 11.07LVLSSGIGNLTTGSVVSFNIYY206 pKa = 9.44IDD208 pKa = 3.67YY209 pKa = 10.97DD210 pKa = 3.26IDD212 pKa = 3.78FEE214 pKa = 4.68NRR216 pKa = 11.84LATPDD221 pKa = 3.44TSSSFIYY228 pKa = 9.68PDD230 pKa = 3.81PGYY233 pKa = 10.91EE234 pKa = 5.4SYY236 pKa = 9.17FTDD239 pKa = 3.64NGDD242 pKa = 4.26GITKK246 pKa = 10.57LSLKK250 pKa = 9.58HH251 pKa = 6.37AEE253 pKa = 4.42GGALVPFHH261 pKa = 6.43QSIPEE266 pKa = 3.78AVYY269 pKa = 10.21EE270 pKa = 4.13FVGEE274 pKa = 4.05SLKK277 pKa = 10.69YY278 pKa = 10.25CKK280 pKa = 9.87TGEE283 pKa = 4.17TSATATGIVTHH294 pKa = 6.41AVRR297 pKa = 11.84IRR299 pKa = 11.84GRR301 pKa = 11.84PDD303 pKa = 2.6GALYY307 pKa = 10.82VFEE310 pKa = 5.45NNTKK314 pKa = 9.85ASAYY318 pKa = 10.39ASTGDD323 pKa = 3.63EE324 pKa = 4.72DD325 pKa = 4.25NCITYY330 pKa = 10.04VAAGPWVSPTGAGFKK345 pKa = 10.36QISSPSALYY354 pKa = 10.75SNRR357 pKa = 11.84LNN359 pKa = 3.59
MM1 pKa = 7.41VKK3 pKa = 9.41TEE5 pKa = 3.87QQRR8 pKa = 11.84AKK10 pKa = 9.79QRR12 pKa = 11.84EE13 pKa = 3.62RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84QRR19 pKa = 11.84QNARR23 pKa = 11.84LRR25 pKa = 11.84KK26 pKa = 9.34EE27 pKa = 4.21MNQEE31 pKa = 3.73APRR34 pKa = 11.84PLPQRR39 pKa = 11.84QRR41 pKa = 11.84GRR43 pKa = 11.84MGGIKK48 pKa = 10.07RR49 pKa = 11.84NCFDD53 pKa = 4.08GNDD56 pKa = 4.42LISSVDD62 pKa = 3.49VPATAKK68 pKa = 10.37SGTVIARR75 pKa = 11.84VDD77 pKa = 3.19ICPSKK82 pKa = 10.96FPGTSLWRR90 pKa = 11.84QAQLYY95 pKa = 9.8ANWKK99 pKa = 7.63PARR102 pKa = 11.84KK103 pKa = 9.27LKK105 pKa = 10.58IVVQPSAGTTTSGSYY120 pKa = 10.48IMGWTMDD127 pKa = 3.73SKK129 pKa = 11.7LDD131 pKa = 3.78IPSGDD136 pKa = 3.29NAVRR140 pKa = 11.84TVATFEE146 pKa = 3.8RR147 pKa = 11.84RR148 pKa = 11.84QTAKK152 pKa = 10.1IYY154 pKa = 9.41EE155 pKa = 4.32KK156 pKa = 10.93KK157 pKa = 10.31EE158 pKa = 3.6LNIPLKK164 pKa = 9.79TVQALLFTDD173 pKa = 3.93SSKK176 pKa = 11.17EE177 pKa = 3.93DD178 pKa = 3.37SDD180 pKa = 3.45QGSFYY185 pKa = 11.07LVLSSGIGNLTTGSVVSFNIYY206 pKa = 9.44IDD208 pKa = 3.67YY209 pKa = 10.97DD210 pKa = 3.26IDD212 pKa = 3.78FEE214 pKa = 4.68NRR216 pKa = 11.84LATPDD221 pKa = 3.44TSSSFIYY228 pKa = 9.68PDD230 pKa = 3.81PGYY233 pKa = 10.91EE234 pKa = 5.4SYY236 pKa = 9.17FTDD239 pKa = 3.64NGDD242 pKa = 4.26GITKK246 pKa = 10.57LSLKK250 pKa = 9.58HH251 pKa = 6.37AEE253 pKa = 4.42GGALVPFHH261 pKa = 6.43QSIPEE266 pKa = 3.78AVYY269 pKa = 10.21EE270 pKa = 4.13FVGEE274 pKa = 4.05SLKK277 pKa = 10.69YY278 pKa = 10.25CKK280 pKa = 9.87TGEE283 pKa = 4.17TSATATGIVTHH294 pKa = 6.41AVRR297 pKa = 11.84IRR299 pKa = 11.84GRR301 pKa = 11.84PDD303 pKa = 2.6GALYY307 pKa = 10.82VFEE310 pKa = 5.45NNTKK314 pKa = 9.85ASAYY318 pKa = 10.39ASTGDD323 pKa = 3.63EE324 pKa = 4.72DD325 pKa = 4.25NCITYY330 pKa = 10.04VAAGPWVSPTGAGFKK345 pKa = 10.36QISSPSALYY354 pKa = 10.75SNRR357 pKa = 11.84LNN359 pKa = 3.59
Molecular weight: 39.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1332 |
359 |
973 |
666.0 |
74.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.381 ± 0.805 | 1.877 ± 0.362 |
6.081 ± 0.242 | 8.859 ± 1.957 |
4.054 ± 0.205 | 7.282 ± 0.245 |
1.276 ± 0.209 | 3.754 ± 0.862 |
7.207 ± 0.644 | 6.757 ± 0.298 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.652 ± 0.123 | 4.129 ± 0.155 |
5.105 ± 0.089 | 3.078 ± 0.654 |
6.456 ± 0.241 | 6.757 ± 1.156 |
6.456 ± 0.637 | 7.808 ± 0.929 |
1.502 ± 0.184 | 3.529 ± 0.308 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
