
Scytalidium sp. 3C
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Scytalidium; unclassified Scytalidium
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
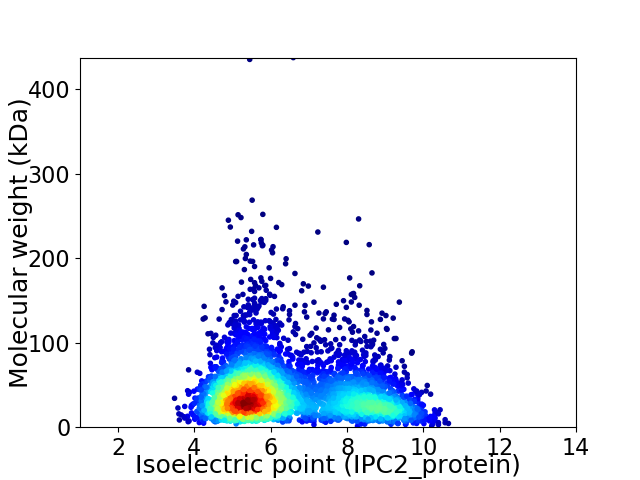
Virtual 2D-PAGE plot for 4330 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A425BTE8|A0A425BTE8_9PEZI Leukotriene A-4 hydrolase homolog OS=Scytalidium sp. 3C OX=2487125 GN=DH86_00001357 PE=3 SV=1
MM1 pKa = 7.23IQGGTQNTVISAIPAAIAQEE21 pKa = 4.17TSLLLGLWASAGQDD35 pKa = 3.36DD36 pKa = 5.14FNNEE40 pKa = 3.17IQALKK45 pKa = 9.4TAIQQFGSQLAPLLVGISVGSEE67 pKa = 3.32DD68 pKa = 5.2LYY70 pKa = 11.01RR71 pKa = 11.84ISPTGVEE78 pKa = 4.26AKK80 pKa = 10.25SGAGANPDD88 pKa = 4.82DD89 pKa = 3.58IVNYY93 pKa = 9.96ISQVRR98 pKa = 11.84DD99 pKa = 3.88AIKK102 pKa = 9.28GTPLSGASIGHH113 pKa = 5.99VDD115 pKa = 3.37TWTAWVNGSNDD126 pKa = 3.41AVINACDD133 pKa = 3.96WIGVDD138 pKa = 4.82AYY140 pKa = 10.08PYY142 pKa = 9.68FQNTMSNGIDD152 pKa = 3.2QGASLFKK159 pKa = 10.25SAYY162 pKa = 10.08SATQQAANGKK172 pKa = 7.48PVWITEE178 pKa = 4.34TGWPVSGSTEE188 pKa = 3.98NLGVPSLQNAKK199 pKa = 9.59TYY201 pKa = 9.17WDD203 pKa = 3.51EE204 pKa = 4.28VGCDD208 pKa = 3.47FAFGKK213 pKa = 10.36INTWWYY219 pKa = 8.64TLQDD223 pKa = 3.99SQPTTPNPSFGVVGSTLSTTPLYY246 pKa = 10.78DD247 pKa = 4.16LSCSSSNSTSSSSSSASSASASATQSSSGASTTQGSGGVGAGSGSGSGSSATTSAAAGGSSSAPSSVAGGNASSAVAAASSGVATGGAGLSPNQANGNGVPPASTGNGTSSGSAAGGSTSGSGASNSTGVASGGSTNSTSSGSGSGSGSGSGSGTSTGSSGSSSSSPITSANSASLVSFPVMGSALVGFAAVVAALL443 pKa = 3.79
MM1 pKa = 7.23IQGGTQNTVISAIPAAIAQEE21 pKa = 4.17TSLLLGLWASAGQDD35 pKa = 3.36DD36 pKa = 5.14FNNEE40 pKa = 3.17IQALKK45 pKa = 9.4TAIQQFGSQLAPLLVGISVGSEE67 pKa = 3.32DD68 pKa = 5.2LYY70 pKa = 11.01RR71 pKa = 11.84ISPTGVEE78 pKa = 4.26AKK80 pKa = 10.25SGAGANPDD88 pKa = 4.82DD89 pKa = 3.58IVNYY93 pKa = 9.96ISQVRR98 pKa = 11.84DD99 pKa = 3.88AIKK102 pKa = 9.28GTPLSGASIGHH113 pKa = 5.99VDD115 pKa = 3.37TWTAWVNGSNDD126 pKa = 3.41AVINACDD133 pKa = 3.96WIGVDD138 pKa = 4.82AYY140 pKa = 10.08PYY142 pKa = 9.68FQNTMSNGIDD152 pKa = 3.2QGASLFKK159 pKa = 10.25SAYY162 pKa = 10.08SATQQAANGKK172 pKa = 7.48PVWITEE178 pKa = 4.34TGWPVSGSTEE188 pKa = 3.98NLGVPSLQNAKK199 pKa = 9.59TYY201 pKa = 9.17WDD203 pKa = 3.51EE204 pKa = 4.28VGCDD208 pKa = 3.47FAFGKK213 pKa = 10.36INTWWYY219 pKa = 8.64TLQDD223 pKa = 3.99SQPTTPNPSFGVVGSTLSTTPLYY246 pKa = 10.78DD247 pKa = 4.16LSCSSSNSTSSSSSSASSASASATQSSSGASTTQGSGGVGAGSGSGSGSSATTSAAAGGSSSAPSSVAGGNASSAVAAASSGVATGGAGLSPNQANGNGVPPASTGNGTSSGSAAGGSTSGSGASNSTGVASGGSTNSTSSGSGSGSGSGSGSGTSTGSSGSSSSSPITSANSASLVSFPVMGSALVGFAAVVAALL443 pKa = 3.79
Molecular weight: 42.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A425BVE2|A0A425BVE2_9PEZI Chitin synthase (Fragment) OS=Scytalidium sp. 3C OX=2487125 GN=DH86_00002431 PE=3 SV=1
KKK2 pKa = 9.89RR3 pKa = 11.84CDDD6 pKa = 2.93KKK8 pKa = 8.82PCSRR12 pKa = 11.84CTRR15 pKa = 11.84LDDD18 pKa = 3.61DDD20 pKa = 3.9ARR22 pKa = 11.84LRR24 pKa = 11.84SHHH27 pKa = 6.57RR28 pKa = 11.84HHH30 pKa = 5.41RR31 pKa = 11.84PANNTSGQDDD41 pKa = 3.59EEE43 pKa = 4.54VEEE46 pKa = 4.21RR47 pKa = 11.84KKK49 pKa = 9.46KK50 pKa = 9.36YY51 pKa = 9.64AVTVRR56 pKa = 11.84RR57 pKa = 11.84DDD59 pKa = 3.17TGCDDD64 pKa = 2.67CKKK67 pKa = 10.37KK68 pKa = 10.37NVTRR72 pKa = 11.84KKK74 pKa = 9.11RR75 pKa = 11.84SAQIVEEE82 pKa = 3.93DD83 pKa = 4.3IYYY86 pKa = 10.12HH87 pKa = 5.32SEEE90 pKa = 4.33FVLPRR95 pKa = 11.84LRR97 pKa = 11.84SHHH100 pKa = 7.52PPLLYYY106 pKa = 10.22QHHH109 pKa = 7.0PPQMPSFRR117 pKa = 11.84SRR119 pKa = 11.84PNQTQLLCPKKK130 pKa = 10.01RR131 pKa = 11.84GPEEE135 pKa = 3.67AVLRR139 pKa = 11.84IQGFSSDDD147 pKa = 3.04HH148 pKa = 5.92LYYY151 pKa = 10.92NGFRR155 pKa = 11.84DDD157 pKa = 3.74DD158 pKa = 5.24LYYY161 pKa = 10.33HHH163 pKa = 7.18HH164 pKa = 7.41RR165 pKa = 11.84PQQISWSLLRR175 pKa = 11.84TQVMMMEEE183 pKa = 4.8VAHHH187 pKa = 6.28LLSGLYYY194 pKa = 9.04VNLLAKKK201 pKa = 9.63LANNRR206 pKa = 11.84IGYYY210 pKa = 10.07STP
KKK2 pKa = 9.89RR3 pKa = 11.84CDDD6 pKa = 2.93KKK8 pKa = 8.82PCSRR12 pKa = 11.84CTRR15 pKa = 11.84LDDD18 pKa = 3.61DDD20 pKa = 3.9ARR22 pKa = 11.84LRR24 pKa = 11.84SHHH27 pKa = 6.57RR28 pKa = 11.84HHH30 pKa = 5.41RR31 pKa = 11.84PANNTSGQDDD41 pKa = 3.59EEE43 pKa = 4.54VEEE46 pKa = 4.21RR47 pKa = 11.84KKK49 pKa = 9.46KK50 pKa = 9.36YY51 pKa = 9.64AVTVRR56 pKa = 11.84RR57 pKa = 11.84DDD59 pKa = 3.17TGCDDD64 pKa = 2.67CKKK67 pKa = 10.37KK68 pKa = 10.37NVTRR72 pKa = 11.84KKK74 pKa = 9.11RR75 pKa = 11.84SAQIVEEE82 pKa = 3.93DD83 pKa = 4.3IYYY86 pKa = 10.12HH87 pKa = 5.32SEEE90 pKa = 4.33FVLPRR95 pKa = 11.84LRR97 pKa = 11.84SHHH100 pKa = 7.52PPLLYYY106 pKa = 10.22QHHH109 pKa = 7.0PPQMPSFRR117 pKa = 11.84SRR119 pKa = 11.84PNQTQLLCPKKK130 pKa = 10.01RR131 pKa = 11.84GPEEE135 pKa = 3.67AVLRR139 pKa = 11.84IQGFSSDDD147 pKa = 3.04HH148 pKa = 5.92LYYY151 pKa = 10.92NGFRR155 pKa = 11.84DDD157 pKa = 3.74DD158 pKa = 5.24LYYY161 pKa = 10.33HHH163 pKa = 7.18HH164 pKa = 7.41RR165 pKa = 11.84PQQISWSLLRR175 pKa = 11.84TQVMMMEEE183 pKa = 4.8VAHHH187 pKa = 6.28LLSGLYYY194 pKa = 9.04VNLLAKKK201 pKa = 9.63LANNRR206 pKa = 11.84IGYYY210 pKa = 10.07STP
Molecular weight: 24.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1786973 |
11 |
3850 |
412.7 |
45.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.675 ± 0.034 | 1.035 ± 0.013 |
5.554 ± 0.029 | 6.471 ± 0.041 |
3.688 ± 0.025 | 7.086 ± 0.036 |
2.186 ± 0.017 | 5.181 ± 0.027 |
5.345 ± 0.036 | 8.667 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.176 ± 0.015 | 3.9 ± 0.022 |
5.966 ± 0.041 | 3.985 ± 0.03 |
5.683 ± 0.034 | 8.133 ± 0.048 |
5.968 ± 0.035 | 6.103 ± 0.031 |
1.334 ± 0.015 | 2.858 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
