
Clostridium sp. W14A
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; unclassified Clostridium
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
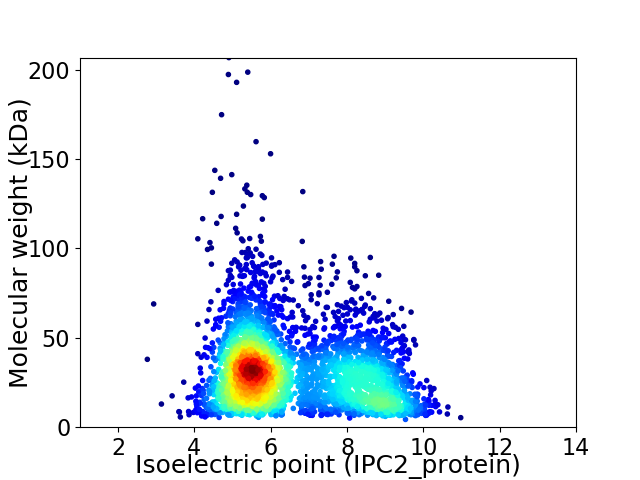
Virtual 2D-PAGE plot for 3645 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C0BMT5|A0A1C0BMT5_9CLOT RNA polymerase subunit sigma-70 OS=Clostridium sp. W14A OX=1849176 GN=A7X67_04595 PE=4 SV=1
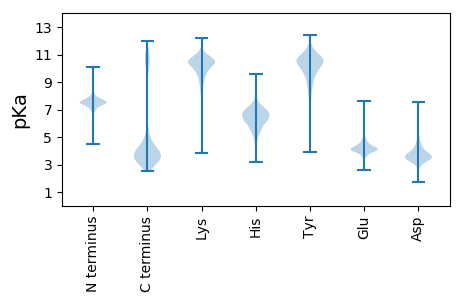
MM1 pKa = 7.62AANIYY6 pKa = 10.0ILSACDD12 pKa = 2.79AWAEE16 pKa = 4.01HH17 pKa = 5.96SSMRR21 pKa = 11.84ILGVTTDD28 pKa = 3.55EE29 pKa = 4.11NMLYY33 pKa = 11.23AMLAAKK39 pKa = 10.13IKK41 pKa = 10.56AGDD44 pKa = 3.65MEE46 pKa = 4.72YY47 pKa = 10.88DD48 pKa = 3.55GSGEE52 pKa = 4.07NAWSKK57 pKa = 9.63FQNDD61 pKa = 4.22FKK63 pKa = 11.57NGDD66 pKa = 3.36INFNKK71 pKa = 10.03LKK73 pKa = 10.59YY74 pKa = 10.6GFVQTYY80 pKa = 9.5EE81 pKa = 4.16DD82 pKa = 3.72MQITEE87 pKa = 5.39PISLAQFPEE96 pKa = 4.55AGEE99 pKa = 4.24VYY101 pKa = 10.77EE102 pKa = 5.18EE103 pKa = 3.76ITGAKK108 pKa = 9.09VRR110 pKa = 11.84ADD112 pKa = 3.7MEE114 pKa = 4.55RR115 pKa = 11.84LGLDD119 pKa = 3.11HH120 pKa = 7.41RR121 pKa = 11.84SLVYY125 pKa = 10.54SVVEE129 pKa = 3.77VHH131 pKa = 6.41TDD133 pKa = 2.98SGDD136 pKa = 3.22TSFYY140 pKa = 10.43MPGICDD146 pKa = 3.3RR147 pKa = 11.84DD148 pKa = 3.81SLEE151 pKa = 4.55EE152 pKa = 4.14NDD154 pKa = 5.71DD155 pKa = 4.02YY156 pKa = 11.94LDD158 pKa = 4.66LMDD161 pKa = 5.33GADD164 pKa = 3.75DD165 pKa = 4.17TEE167 pKa = 4.62VDD169 pKa = 3.9VSVSSYY175 pKa = 11.65SLGTGEE181 pKa = 5.23SEE183 pKa = 4.34YY184 pKa = 10.53PDD186 pKa = 3.71EE187 pKa = 4.41EE188 pKa = 4.27EE189 pKa = 3.95IAIIEE194 pKa = 4.3QYY196 pKa = 10.37TDD198 pKa = 3.63EE199 pKa = 5.4LDD201 pKa = 3.39EE202 pKa = 5.08EE203 pKa = 4.63YY204 pKa = 10.76GIDD207 pKa = 4.5PIQSDD212 pKa = 4.03SFSFEE217 pKa = 3.92YY218 pKa = 10.43EE219 pKa = 4.0AEE221 pKa = 4.23QEE223 pKa = 4.44CC224 pKa = 4.91
MM1 pKa = 7.62AANIYY6 pKa = 10.0ILSACDD12 pKa = 2.79AWAEE16 pKa = 4.01HH17 pKa = 5.96SSMRR21 pKa = 11.84ILGVTTDD28 pKa = 3.55EE29 pKa = 4.11NMLYY33 pKa = 11.23AMLAAKK39 pKa = 10.13IKK41 pKa = 10.56AGDD44 pKa = 3.65MEE46 pKa = 4.72YY47 pKa = 10.88DD48 pKa = 3.55GSGEE52 pKa = 4.07NAWSKK57 pKa = 9.63FQNDD61 pKa = 4.22FKK63 pKa = 11.57NGDD66 pKa = 3.36INFNKK71 pKa = 10.03LKK73 pKa = 10.59YY74 pKa = 10.6GFVQTYY80 pKa = 9.5EE81 pKa = 4.16DD82 pKa = 3.72MQITEE87 pKa = 5.39PISLAQFPEE96 pKa = 4.55AGEE99 pKa = 4.24VYY101 pKa = 10.77EE102 pKa = 5.18EE103 pKa = 3.76ITGAKK108 pKa = 9.09VRR110 pKa = 11.84ADD112 pKa = 3.7MEE114 pKa = 4.55RR115 pKa = 11.84LGLDD119 pKa = 3.11HH120 pKa = 7.41RR121 pKa = 11.84SLVYY125 pKa = 10.54SVVEE129 pKa = 3.77VHH131 pKa = 6.41TDD133 pKa = 2.98SGDD136 pKa = 3.22TSFYY140 pKa = 10.43MPGICDD146 pKa = 3.3RR147 pKa = 11.84DD148 pKa = 3.81SLEE151 pKa = 4.55EE152 pKa = 4.14NDD154 pKa = 5.71DD155 pKa = 4.02YY156 pKa = 11.94LDD158 pKa = 4.66LMDD161 pKa = 5.33GADD164 pKa = 3.75DD165 pKa = 4.17TEE167 pKa = 4.62VDD169 pKa = 3.9VSVSSYY175 pKa = 11.65SLGTGEE181 pKa = 5.23SEE183 pKa = 4.34YY184 pKa = 10.53PDD186 pKa = 3.71EE187 pKa = 4.41EE188 pKa = 4.27EE189 pKa = 3.95IAIIEE194 pKa = 4.3QYY196 pKa = 10.37TDD198 pKa = 3.63EE199 pKa = 5.4LDD201 pKa = 3.39EE202 pKa = 5.08EE203 pKa = 4.63YY204 pKa = 10.76GIDD207 pKa = 4.5PIQSDD212 pKa = 4.03SFSFEE217 pKa = 3.92YY218 pKa = 10.43EE219 pKa = 4.0AEE221 pKa = 4.23QEE223 pKa = 4.44CC224 pKa = 4.91
Molecular weight: 25.21 kDa
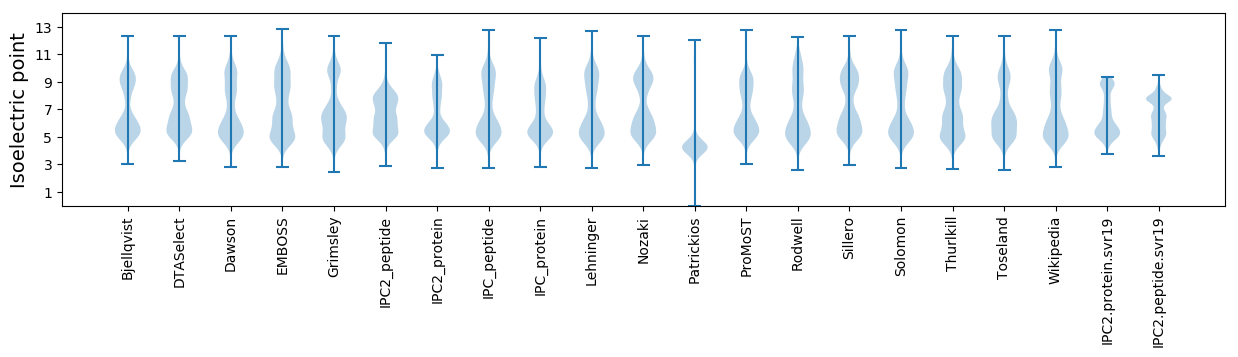
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C0BLY7|A0A1C0BLY7_9CLOT Carboxymuconolactone decarboxylase OS=Clostridium sp. W14A OX=1849176 GN=A7X67_01005 PE=4 SV=1
MM1 pKa = 7.19KK2 pKa = 10.14RR3 pKa = 11.84KK4 pKa = 8.97HH5 pKa = 6.15KK6 pKa = 10.47KK7 pKa = 9.82YY8 pKa = 10.46LVEE11 pKa = 4.32FWKK14 pKa = 10.7IKK16 pKa = 10.28DD17 pKa = 3.6YY18 pKa = 11.85NEE20 pKa = 3.62IARR23 pKa = 11.84QIKK26 pKa = 10.11SAMQRR31 pKa = 11.84QSVSPLCPLHH41 pKa = 6.56TEE43 pKa = 3.92HH44 pKa = 6.54QPPYY48 pKa = 7.81TTEE51 pKa = 4.02FTAHH55 pKa = 6.4CRR57 pKa = 11.84NHH59 pKa = 4.79YY60 pKa = 9.33TSFFRR65 pKa = 11.84FCTEE69 pKa = 3.33GRR71 pKa = 11.84KK72 pKa = 8.37MVHH75 pKa = 5.76SLIARR80 pKa = 11.84APVYY84 pKa = 10.95GMTFFRR90 pKa = 11.84TGAFCFLAGNGCRR103 pKa = 11.84ASQRR107 pKa = 11.84VNSSAGTRR115 pKa = 11.84RR116 pKa = 11.84HH117 pKa = 4.67VPRR120 pKa = 11.84RR121 pKa = 11.84QNN123 pKa = 2.86
MM1 pKa = 7.19KK2 pKa = 10.14RR3 pKa = 11.84KK4 pKa = 8.97HH5 pKa = 6.15KK6 pKa = 10.47KK7 pKa = 9.82YY8 pKa = 10.46LVEE11 pKa = 4.32FWKK14 pKa = 10.7IKK16 pKa = 10.28DD17 pKa = 3.6YY18 pKa = 11.85NEE20 pKa = 3.62IARR23 pKa = 11.84QIKK26 pKa = 10.11SAMQRR31 pKa = 11.84QSVSPLCPLHH41 pKa = 6.56TEE43 pKa = 3.92HH44 pKa = 6.54QPPYY48 pKa = 7.81TTEE51 pKa = 4.02FTAHH55 pKa = 6.4CRR57 pKa = 11.84NHH59 pKa = 4.79YY60 pKa = 9.33TSFFRR65 pKa = 11.84FCTEE69 pKa = 3.33GRR71 pKa = 11.84KK72 pKa = 8.37MVHH75 pKa = 5.76SLIARR80 pKa = 11.84APVYY84 pKa = 10.95GMTFFRR90 pKa = 11.84TGAFCFLAGNGCRR103 pKa = 11.84ASQRR107 pKa = 11.84VNSSAGTRR115 pKa = 11.84RR116 pKa = 11.84HH117 pKa = 4.67VPRR120 pKa = 11.84RR121 pKa = 11.84QNN123 pKa = 2.86
Molecular weight: 14.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1085271 |
37 |
1975 |
297.7 |
32.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.521 ± 0.044 | 1.592 ± 0.018 |
5.333 ± 0.031 | 6.441 ± 0.046 |
4.26 ± 0.032 | 7.648 ± 0.048 |
1.685 ± 0.017 | 6.508 ± 0.036 |
5.818 ± 0.035 | 9.643 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.712 ± 0.019 | 3.954 ± 0.024 |
3.959 ± 0.03 | 3.302 ± 0.023 |
5.38 ± 0.04 | 6.644 ± 0.037 |
5.342 ± 0.037 | 6.904 ± 0.029 |
0.888 ± 0.014 | 3.466 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
