
Amasya cherry disease-associated mycovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus; Cherry chlorotic rusty spot associated partitivirus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
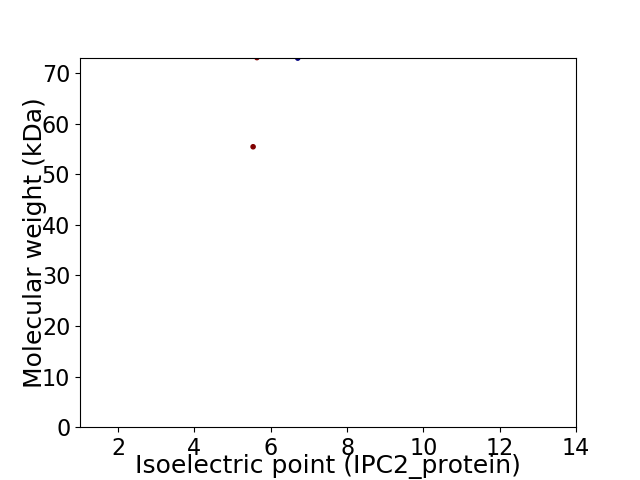
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q65A76|Q65A76_9VIRU Putative coat protein OS=Amasya cherry disease-associated mycovirus OX=284689 GN=CP PE=4 SV=1
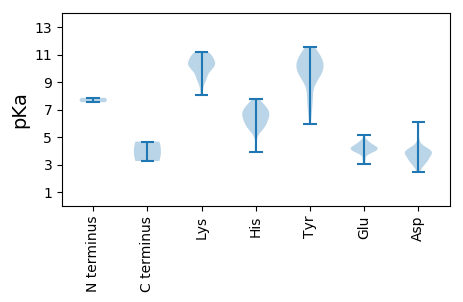
MM1 pKa = 7.53SAPATTGSTTSTAAVPATEE20 pKa = 4.05QTAPATVAAPPTAPATDD37 pKa = 3.48VKK39 pKa = 11.1YY40 pKa = 10.55VAPAKK45 pKa = 9.2KK46 pKa = 9.62QKK48 pKa = 10.55SFAPRR53 pKa = 11.84EE54 pKa = 4.19PTASSAGPKK63 pKa = 10.24NPGLSMMLSGVSDD76 pKa = 3.86LPFFGIKK83 pKa = 10.26HH84 pKa = 5.98NDD86 pKa = 2.78ISYY89 pKa = 9.91VVPDD93 pKa = 4.08TTQLFYY99 pKa = 11.31VLSIMDD105 pKa = 3.71TQMVRR110 pKa = 11.84TKK112 pKa = 10.62RR113 pKa = 11.84FTDD116 pKa = 4.23ANPDD120 pKa = 2.97WHH122 pKa = 6.9PFVSQLYY129 pKa = 8.84IAVLFYY135 pKa = 11.06YY136 pKa = 10.09QVLKK140 pKa = 10.37NQSHH144 pKa = 6.61GGMITNDD151 pKa = 2.61QRR153 pKa = 11.84LFVEE157 pKa = 5.12FLDD160 pKa = 3.95SQFKK164 pKa = 10.92AEE166 pKa = 4.29HH167 pKa = 6.18LKK169 pKa = 10.47IPGPIAIFFQSLAANAGPNEE189 pKa = 4.06NFGNLVFGIPNAHH202 pKa = 7.6DD203 pKa = 3.67INCTSFLWQDD213 pKa = 3.2KK214 pKa = 9.08VHH216 pKa = 6.92TILPNVIFILDD227 pKa = 3.29QFMRR231 pKa = 11.84LISLISPVNSGPVQANASHH250 pKa = 7.08TDD252 pKa = 3.31TVYY255 pKa = 8.22TTIFGAPASKK265 pKa = 11.03DD266 pKa = 3.3EE267 pKa = 4.14ATRR270 pKa = 11.84FAMLTPSARR279 pKa = 11.84SDD281 pKa = 3.44FQTTVGLLNGLATSSNVWRR300 pKa = 11.84NTLPFANDD308 pKa = 3.2GDD310 pKa = 4.26SQYY313 pKa = 11.52VISDD317 pKa = 3.93DD318 pKa = 6.08DD319 pKa = 3.84DD320 pKa = 4.13TLDD323 pKa = 3.92LDD325 pKa = 3.73QVFGFRR331 pKa = 11.84GIGNHH336 pKa = 6.59ADD338 pKa = 3.71RR339 pKa = 11.84PYY341 pKa = 10.87GWFAQVIRR349 pKa = 11.84VMQPYY354 pKa = 10.75SDD356 pKa = 4.09FFKK359 pKa = 10.59DD360 pKa = 3.71TVSLGSVTTTGTGISYY376 pKa = 10.35VRR378 pKa = 11.84TKK380 pKa = 10.91YY381 pKa = 10.57VATRR385 pKa = 11.84QNKK388 pKa = 9.37NILVHH393 pKa = 6.68AVTTRR398 pKa = 11.84KK399 pKa = 9.29VRR401 pKa = 11.84YY402 pKa = 6.57QTGSTLRR409 pKa = 11.84YY410 pKa = 9.43DD411 pKa = 3.22IPEE414 pKa = 3.93FTGITTIHH422 pKa = 5.81LHH424 pKa = 5.56SEE426 pKa = 4.04EE427 pKa = 4.21FLDD430 pKa = 5.74LVTEE434 pKa = 4.06QAGILTQLNSDD445 pKa = 3.05WTDD448 pKa = 2.76INKK451 pKa = 10.19DD452 pKa = 3.4VTDD455 pKa = 3.82TSNPDD460 pKa = 3.27FKK462 pKa = 10.25STHH465 pKa = 5.7VGPIFSIPDD474 pKa = 3.27SRR476 pKa = 11.84RR477 pKa = 11.84TSQYY481 pKa = 10.79NVSNTIAPLISGYY494 pKa = 9.37YY495 pKa = 7.17HH496 pKa = 6.47TPSALRR502 pKa = 11.84FEE504 pKa = 4.67
MM1 pKa = 7.53SAPATTGSTTSTAAVPATEE20 pKa = 4.05QTAPATVAAPPTAPATDD37 pKa = 3.48VKK39 pKa = 11.1YY40 pKa = 10.55VAPAKK45 pKa = 9.2KK46 pKa = 9.62QKK48 pKa = 10.55SFAPRR53 pKa = 11.84EE54 pKa = 4.19PTASSAGPKK63 pKa = 10.24NPGLSMMLSGVSDD76 pKa = 3.86LPFFGIKK83 pKa = 10.26HH84 pKa = 5.98NDD86 pKa = 2.78ISYY89 pKa = 9.91VVPDD93 pKa = 4.08TTQLFYY99 pKa = 11.31VLSIMDD105 pKa = 3.71TQMVRR110 pKa = 11.84TKK112 pKa = 10.62RR113 pKa = 11.84FTDD116 pKa = 4.23ANPDD120 pKa = 2.97WHH122 pKa = 6.9PFVSQLYY129 pKa = 8.84IAVLFYY135 pKa = 11.06YY136 pKa = 10.09QVLKK140 pKa = 10.37NQSHH144 pKa = 6.61GGMITNDD151 pKa = 2.61QRR153 pKa = 11.84LFVEE157 pKa = 5.12FLDD160 pKa = 3.95SQFKK164 pKa = 10.92AEE166 pKa = 4.29HH167 pKa = 6.18LKK169 pKa = 10.47IPGPIAIFFQSLAANAGPNEE189 pKa = 4.06NFGNLVFGIPNAHH202 pKa = 7.6DD203 pKa = 3.67INCTSFLWQDD213 pKa = 3.2KK214 pKa = 9.08VHH216 pKa = 6.92TILPNVIFILDD227 pKa = 3.29QFMRR231 pKa = 11.84LISLISPVNSGPVQANASHH250 pKa = 7.08TDD252 pKa = 3.31TVYY255 pKa = 8.22TTIFGAPASKK265 pKa = 11.03DD266 pKa = 3.3EE267 pKa = 4.14ATRR270 pKa = 11.84FAMLTPSARR279 pKa = 11.84SDD281 pKa = 3.44FQTTVGLLNGLATSSNVWRR300 pKa = 11.84NTLPFANDD308 pKa = 3.2GDD310 pKa = 4.26SQYY313 pKa = 11.52VISDD317 pKa = 3.93DD318 pKa = 6.08DD319 pKa = 3.84DD320 pKa = 4.13TLDD323 pKa = 3.92LDD325 pKa = 3.73QVFGFRR331 pKa = 11.84GIGNHH336 pKa = 6.59ADD338 pKa = 3.71RR339 pKa = 11.84PYY341 pKa = 10.87GWFAQVIRR349 pKa = 11.84VMQPYY354 pKa = 10.75SDD356 pKa = 4.09FFKK359 pKa = 10.59DD360 pKa = 3.71TVSLGSVTTTGTGISYY376 pKa = 10.35VRR378 pKa = 11.84TKK380 pKa = 10.91YY381 pKa = 10.57VATRR385 pKa = 11.84QNKK388 pKa = 9.37NILVHH393 pKa = 6.68AVTTRR398 pKa = 11.84KK399 pKa = 9.29VRR401 pKa = 11.84YY402 pKa = 6.57QTGSTLRR409 pKa = 11.84YY410 pKa = 9.43DD411 pKa = 3.22IPEE414 pKa = 3.93FTGITTIHH422 pKa = 5.81LHH424 pKa = 5.56SEE426 pKa = 4.04EE427 pKa = 4.21FLDD430 pKa = 5.74LVTEE434 pKa = 4.06QAGILTQLNSDD445 pKa = 3.05WTDD448 pKa = 2.76INKK451 pKa = 10.19DD452 pKa = 3.4VTDD455 pKa = 3.82TSNPDD460 pKa = 3.27FKK462 pKa = 10.25STHH465 pKa = 5.7VGPIFSIPDD474 pKa = 3.27SRR476 pKa = 11.84RR477 pKa = 11.84TSQYY481 pKa = 10.79NVSNTIAPLISGYY494 pKa = 9.37YY495 pKa = 7.17HH496 pKa = 6.47TPSALRR502 pKa = 11.84FEE504 pKa = 4.67
Molecular weight: 55.46 kDa
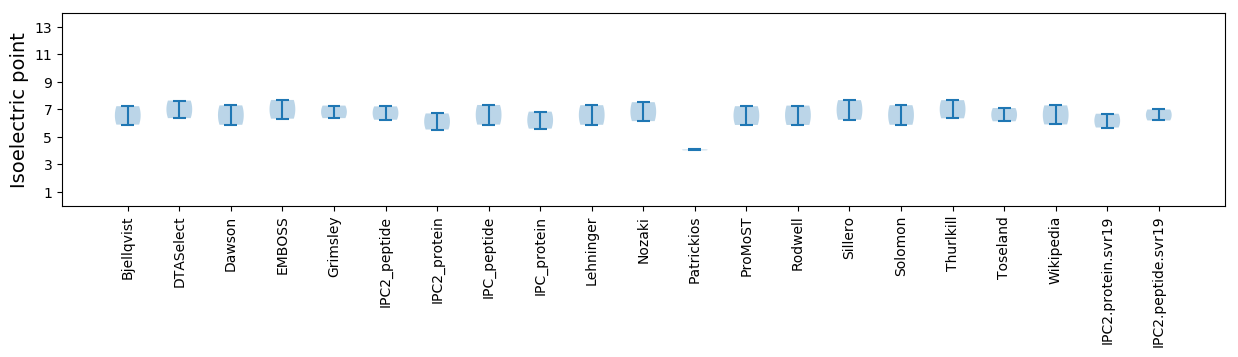
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65A76|Q65A76_9VIRU Putative coat protein OS=Amasya cherry disease-associated mycovirus OX=284689 GN=CP PE=4 SV=1
MM1 pKa = 7.85DD2 pKa = 5.56HH3 pKa = 6.36LTSLFEE9 pKa = 4.7LFAITPKK16 pKa = 9.25TQNNLQFVGIYY27 pKa = 9.49HH28 pKa = 7.2RR29 pKa = 11.84PPHH32 pKa = 5.77SVRR35 pKa = 11.84ANLRR39 pKa = 11.84NVEE42 pKa = 4.02KK43 pKa = 10.89HH44 pKa = 6.1KK45 pKa = 10.22ITVAHH50 pKa = 6.85AMHH53 pKa = 7.1KK54 pKa = 9.95YY55 pKa = 10.33LYY57 pKa = 8.6PHH59 pKa = 7.31EE60 pKa = 4.72IDD62 pKa = 4.64FVINQMRR69 pKa = 11.84RR70 pKa = 11.84SDD72 pKa = 3.7VTEE75 pKa = 4.03DD76 pKa = 4.22AILADD81 pKa = 4.1FFDD84 pKa = 4.77NNVEE88 pKa = 3.98PLEE91 pKa = 4.37PVLDD95 pKa = 3.67EE96 pKa = 4.21HH97 pKa = 7.36FEE99 pKa = 4.31RR100 pKa = 11.84GLSAMLDD107 pKa = 3.51AFRR110 pKa = 11.84PPQKK114 pKa = 10.17CLPAHH119 pKa = 6.79IYY121 pKa = 9.98DD122 pKa = 4.15VQHH125 pKa = 6.75HH126 pKa = 6.33YY127 pKa = 9.93PYY129 pKa = 10.23KK130 pKa = 8.07WQVNAEE136 pKa = 4.32APFSTDD142 pKa = 3.2SYY144 pKa = 11.38FLANRR149 pKa = 11.84PTFRR153 pKa = 11.84AVFEE157 pKa = 4.34RR158 pKa = 11.84LEE160 pKa = 3.87SLYY163 pKa = 10.24THH165 pKa = 7.79LATDD169 pKa = 2.74WHH171 pKa = 5.95RR172 pKa = 11.84RR173 pKa = 11.84YY174 pKa = 10.71GNKK177 pKa = 8.83TDD179 pKa = 5.12NDD181 pKa = 4.14DD182 pKa = 3.93FMNDD186 pKa = 3.05HH187 pKa = 6.34VPAKK191 pKa = 9.77FGPMKK196 pKa = 9.67EE197 pKa = 4.61TVFSWTHH204 pKa = 5.21RR205 pKa = 11.84WHH207 pKa = 6.89HH208 pKa = 5.56VIKK211 pKa = 11.19SNFTDD216 pKa = 3.39TAGLSKK222 pKa = 10.64DD223 pKa = 3.67YY224 pKa = 10.71YY225 pKa = 10.71FKK227 pKa = 11.1NRR229 pKa = 11.84YY230 pKa = 7.77IFPMLLHH237 pKa = 6.66TKK239 pKa = 8.25TAIVKK244 pKa = 10.22KK245 pKa = 10.36DD246 pKa = 3.95DD247 pKa = 4.07PNKK250 pKa = 9.8MRR252 pKa = 11.84TIWGCSKK259 pKa = 10.15PWIIADD265 pKa = 3.57TMLWWEE271 pKa = 4.0YY272 pKa = 10.05VAYY275 pKa = 10.38AKK277 pKa = 9.55LQPGATPMLWSYY289 pKa = 8.05EE290 pKa = 4.16TFTGGWLRR298 pKa = 11.84LNHH301 pKa = 6.83ALFSSYY307 pKa = 9.7IRR309 pKa = 11.84HH310 pKa = 6.34SYY312 pKa = 7.64ITLDD316 pKa = 3.11WKK318 pKa = 11.11RR319 pKa = 11.84FDD321 pKa = 3.44KK322 pKa = 10.8KK323 pKa = 11.06AYY325 pKa = 9.7FCIIDD330 pKa = 3.92KK331 pKa = 10.84IFDD334 pKa = 3.79GVEE337 pKa = 3.79TFLDD341 pKa = 3.87FDD343 pKa = 3.92NGYY346 pKa = 10.21LPTKK350 pKa = 9.96DD351 pKa = 3.85YY352 pKa = 11.13PDD354 pKa = 3.92TKK356 pKa = 10.44STWTQEE362 pKa = 3.02RR363 pKa = 11.84STRR366 pKa = 11.84LKK368 pKa = 10.86RR369 pKa = 11.84LFDD372 pKa = 3.25WTKK375 pKa = 10.07EE376 pKa = 4.22NFYY379 pKa = 10.19HH380 pKa = 7.28APIVLPNGHH389 pKa = 6.02MYY391 pKa = 9.48VRR393 pKa = 11.84KK394 pKa = 8.98FAGIPSGLFITQLIDD409 pKa = 2.46SWYY412 pKa = 9.4NYY414 pKa = 9.25TMLATILSAMGFDD427 pKa = 3.97PRR429 pKa = 11.84SCIIKK434 pKa = 9.98VQGDD438 pKa = 3.52DD439 pKa = 3.9SIIRR443 pKa = 11.84LSALIPPDD451 pKa = 3.22AHH453 pKa = 7.43DD454 pKa = 4.05SFLTKK459 pKa = 10.32VQEE462 pKa = 3.84LADD465 pKa = 3.93YY466 pKa = 9.34YY467 pKa = 9.39FQSVVSVNKK476 pKa = 10.38SEE478 pKa = 4.66VRR480 pKa = 11.84NEE482 pKa = 3.87LNGCEE487 pKa = 4.1VLSYY491 pKa = 9.85RR492 pKa = 11.84HH493 pKa = 5.67RR494 pKa = 11.84HH495 pKa = 4.39GLPYY499 pKa = 9.63RR500 pKa = 11.84DD501 pKa = 4.09EE502 pKa = 4.56LAMLAQLYY510 pKa = 7.18HH511 pKa = 5.81TKK513 pKa = 10.5ARR515 pKa = 11.84NPSPEE520 pKa = 3.35ITMAQSIGFAYY531 pKa = 10.29ASFGNHH537 pKa = 3.93EE538 pKa = 4.43RR539 pKa = 11.84VRR541 pKa = 11.84LVLHH545 pKa = 7.38DD546 pKa = 2.91IHH548 pKa = 7.1EE549 pKa = 4.48YY550 pKa = 10.99YY551 pKa = 10.61KK552 pKa = 10.99LQGYY556 pKa = 5.98TPNRR560 pKa = 11.84AGLSLVFGNSPDD572 pKa = 4.23LMIPHH577 pKa = 6.13YY578 pKa = 9.49TLDD581 pKa = 3.82HH582 pKa = 6.25FPSLRR587 pKa = 11.84EE588 pKa = 3.4IKK590 pKa = 10.1MFLTNAEE597 pKa = 4.3YY598 pKa = 11.25VNEE601 pKa = 4.22EE602 pKa = 4.36TNSRR606 pKa = 11.84TWPLNHH612 pKa = 6.66FLHH615 pKa = 6.6LPCHH619 pKa = 5.54RR620 pKa = 11.84TT621 pKa = 3.25
MM1 pKa = 7.85DD2 pKa = 5.56HH3 pKa = 6.36LTSLFEE9 pKa = 4.7LFAITPKK16 pKa = 9.25TQNNLQFVGIYY27 pKa = 9.49HH28 pKa = 7.2RR29 pKa = 11.84PPHH32 pKa = 5.77SVRR35 pKa = 11.84ANLRR39 pKa = 11.84NVEE42 pKa = 4.02KK43 pKa = 10.89HH44 pKa = 6.1KK45 pKa = 10.22ITVAHH50 pKa = 6.85AMHH53 pKa = 7.1KK54 pKa = 9.95YY55 pKa = 10.33LYY57 pKa = 8.6PHH59 pKa = 7.31EE60 pKa = 4.72IDD62 pKa = 4.64FVINQMRR69 pKa = 11.84RR70 pKa = 11.84SDD72 pKa = 3.7VTEE75 pKa = 4.03DD76 pKa = 4.22AILADD81 pKa = 4.1FFDD84 pKa = 4.77NNVEE88 pKa = 3.98PLEE91 pKa = 4.37PVLDD95 pKa = 3.67EE96 pKa = 4.21HH97 pKa = 7.36FEE99 pKa = 4.31RR100 pKa = 11.84GLSAMLDD107 pKa = 3.51AFRR110 pKa = 11.84PPQKK114 pKa = 10.17CLPAHH119 pKa = 6.79IYY121 pKa = 9.98DD122 pKa = 4.15VQHH125 pKa = 6.75HH126 pKa = 6.33YY127 pKa = 9.93PYY129 pKa = 10.23KK130 pKa = 8.07WQVNAEE136 pKa = 4.32APFSTDD142 pKa = 3.2SYY144 pKa = 11.38FLANRR149 pKa = 11.84PTFRR153 pKa = 11.84AVFEE157 pKa = 4.34RR158 pKa = 11.84LEE160 pKa = 3.87SLYY163 pKa = 10.24THH165 pKa = 7.79LATDD169 pKa = 2.74WHH171 pKa = 5.95RR172 pKa = 11.84RR173 pKa = 11.84YY174 pKa = 10.71GNKK177 pKa = 8.83TDD179 pKa = 5.12NDD181 pKa = 4.14DD182 pKa = 3.93FMNDD186 pKa = 3.05HH187 pKa = 6.34VPAKK191 pKa = 9.77FGPMKK196 pKa = 9.67EE197 pKa = 4.61TVFSWTHH204 pKa = 5.21RR205 pKa = 11.84WHH207 pKa = 6.89HH208 pKa = 5.56VIKK211 pKa = 11.19SNFTDD216 pKa = 3.39TAGLSKK222 pKa = 10.64DD223 pKa = 3.67YY224 pKa = 10.71YY225 pKa = 10.71FKK227 pKa = 11.1NRR229 pKa = 11.84YY230 pKa = 7.77IFPMLLHH237 pKa = 6.66TKK239 pKa = 8.25TAIVKK244 pKa = 10.22KK245 pKa = 10.36DD246 pKa = 3.95DD247 pKa = 4.07PNKK250 pKa = 9.8MRR252 pKa = 11.84TIWGCSKK259 pKa = 10.15PWIIADD265 pKa = 3.57TMLWWEE271 pKa = 4.0YY272 pKa = 10.05VAYY275 pKa = 10.38AKK277 pKa = 9.55LQPGATPMLWSYY289 pKa = 8.05EE290 pKa = 4.16TFTGGWLRR298 pKa = 11.84LNHH301 pKa = 6.83ALFSSYY307 pKa = 9.7IRR309 pKa = 11.84HH310 pKa = 6.34SYY312 pKa = 7.64ITLDD316 pKa = 3.11WKK318 pKa = 11.11RR319 pKa = 11.84FDD321 pKa = 3.44KK322 pKa = 10.8KK323 pKa = 11.06AYY325 pKa = 9.7FCIIDD330 pKa = 3.92KK331 pKa = 10.84IFDD334 pKa = 3.79GVEE337 pKa = 3.79TFLDD341 pKa = 3.87FDD343 pKa = 3.92NGYY346 pKa = 10.21LPTKK350 pKa = 9.96DD351 pKa = 3.85YY352 pKa = 11.13PDD354 pKa = 3.92TKK356 pKa = 10.44STWTQEE362 pKa = 3.02RR363 pKa = 11.84STRR366 pKa = 11.84LKK368 pKa = 10.86RR369 pKa = 11.84LFDD372 pKa = 3.25WTKK375 pKa = 10.07EE376 pKa = 4.22NFYY379 pKa = 10.19HH380 pKa = 7.28APIVLPNGHH389 pKa = 6.02MYY391 pKa = 9.48VRR393 pKa = 11.84KK394 pKa = 8.98FAGIPSGLFITQLIDD409 pKa = 2.46SWYY412 pKa = 9.4NYY414 pKa = 9.25TMLATILSAMGFDD427 pKa = 3.97PRR429 pKa = 11.84SCIIKK434 pKa = 9.98VQGDD438 pKa = 3.52DD439 pKa = 3.9SIIRR443 pKa = 11.84LSALIPPDD451 pKa = 3.22AHH453 pKa = 7.43DD454 pKa = 4.05SFLTKK459 pKa = 10.32VQEE462 pKa = 3.84LADD465 pKa = 3.93YY466 pKa = 9.34YY467 pKa = 9.39FQSVVSVNKK476 pKa = 10.38SEE478 pKa = 4.66VRR480 pKa = 11.84NEE482 pKa = 3.87LNGCEE487 pKa = 4.1VLSYY491 pKa = 9.85RR492 pKa = 11.84HH493 pKa = 5.67RR494 pKa = 11.84HH495 pKa = 4.39GLPYY499 pKa = 9.63RR500 pKa = 11.84DD501 pKa = 4.09EE502 pKa = 4.56LAMLAQLYY510 pKa = 7.18HH511 pKa = 5.81TKK513 pKa = 10.5ARR515 pKa = 11.84NPSPEE520 pKa = 3.35ITMAQSIGFAYY531 pKa = 10.29ASFGNHH537 pKa = 3.93EE538 pKa = 4.43RR539 pKa = 11.84VRR541 pKa = 11.84LVLHH545 pKa = 7.38DD546 pKa = 2.91IHH548 pKa = 7.1EE549 pKa = 4.48YY550 pKa = 10.99YY551 pKa = 10.61KK552 pKa = 10.99LQGYY556 pKa = 5.98TPNRR560 pKa = 11.84AGLSLVFGNSPDD572 pKa = 4.23LMIPHH577 pKa = 6.13YY578 pKa = 9.49TLDD581 pKa = 3.82HH582 pKa = 6.25FPSLRR587 pKa = 11.84EE588 pKa = 3.4IKK590 pKa = 10.1MFLTNAEE597 pKa = 4.3YY598 pKa = 11.25VNEE601 pKa = 4.22EE602 pKa = 4.36TNSRR606 pKa = 11.84TWPLNHH612 pKa = 6.66FLHH615 pKa = 6.6LPCHH619 pKa = 5.54RR620 pKa = 11.84TT621 pKa = 3.25
Molecular weight: 72.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1125 |
504 |
621 |
562.5 |
64.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.933 ± 0.673 | 0.622 ± 0.284 |
6.578 ± 0.113 | 3.556 ± 0.921 |
6.133 ± 0.012 | 4.533 ± 0.686 |
4.178 ± 1.073 | 5.6 ± 0.236 |
4.533 ± 0.646 | 8.178 ± 0.828 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.311 ± 0.353 | 5.067 ± 0.062 |
6.133 ± 0.278 | 3.378 ± 0.796 |
4.8 ± 0.691 | 6.844 ± 0.999 |
8.356 ± 1.45 | 5.867 ± 0.856 |
1.778 ± 0.527 | 4.622 ± 0.838 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
