
Etheostoma spectabile (orangethroat darter)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala;
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
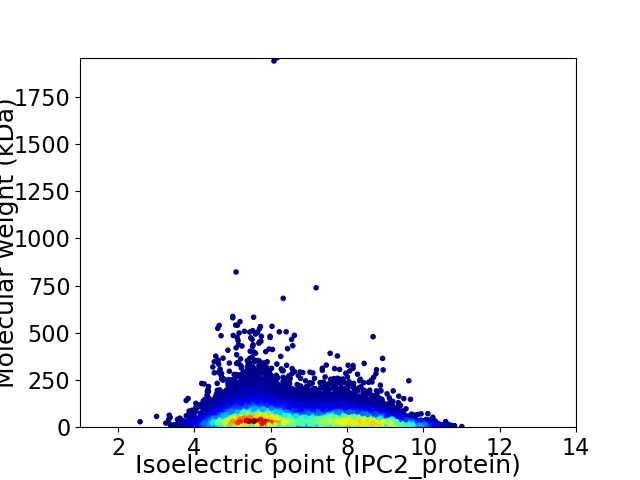
Virtual 2D-PAGE plot for 18371 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
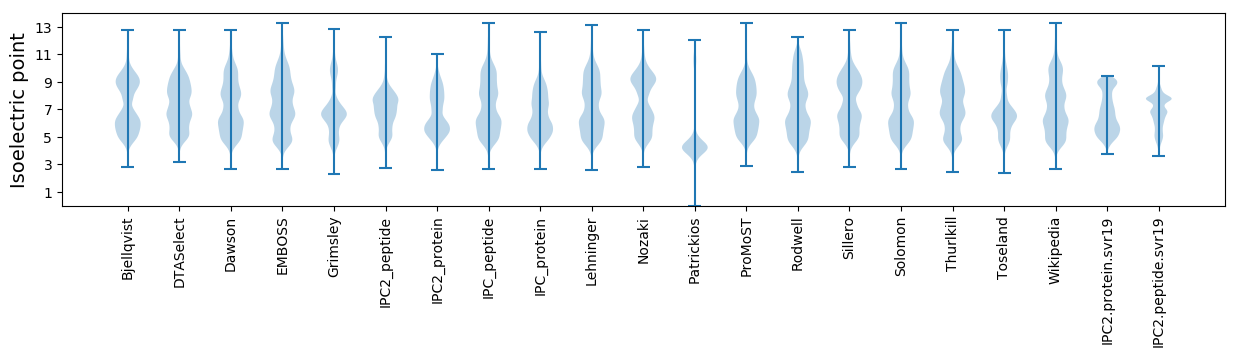
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J5DIY8|A0A5J5DIY8_9PERO Uncharacterized protein OS=Etheostoma spectabile OX=54343 GN=FQN60_009397 PE=4 SV=1
MM1 pKa = 7.73DD2 pKa = 5.74HH3 pKa = 6.77SPSIITQVTRR13 pKa = 11.84DD14 pKa = 3.57EE15 pKa = 4.6EE16 pKa = 5.03DD17 pKa = 3.16NATCRR22 pKa = 11.84EE23 pKa = 4.08DD24 pKa = 3.4GANEE28 pKa = 3.7VSTSKK33 pKa = 10.59KK34 pKa = 8.14PRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GLLDD42 pKa = 3.85SLFCCLCRR50 pKa = 11.84KK51 pKa = 9.23EE52 pKa = 5.13SEE54 pKa = 4.28PPPLKK59 pKa = 10.92NNAPLLVEE67 pKa = 4.34EE68 pKa = 5.1NGTLSKK74 pKa = 10.43QCKK77 pKa = 9.54PSDD80 pKa = 3.63QQFEE84 pKa = 4.41SSDD87 pKa = 3.87FEE89 pKa = 5.82PDD91 pKa = 3.17TLAVNHH97 pKa = 7.1DD98 pKa = 3.83GFGQCEE104 pKa = 4.05MTDD107 pKa = 4.4FIPEE111 pKa = 4.35CTDD114 pKa = 3.35HH115 pKa = 8.14LDD117 pKa = 3.65FLQHH121 pKa = 6.31YY122 pKa = 7.21EE123 pKa = 4.09PCDD126 pKa = 3.53PQCEE130 pKa = 4.28YY131 pKa = 11.13FEE133 pKa = 5.97PDD135 pKa = 3.85TSTEE139 pKa = 3.88DD140 pKa = 3.35CKK142 pKa = 10.88QCEE145 pKa = 4.26RR146 pKa = 11.84TGFTPNVSDD155 pKa = 5.82SEE157 pKa = 4.57DD158 pKa = 3.65LLDD161 pKa = 5.15CGTEE165 pKa = 3.62HH166 pKa = 7.47FEE168 pKa = 5.02YY169 pKa = 10.83DD170 pKa = 3.31EE171 pKa = 4.61CTNDD175 pKa = 5.61DD176 pKa = 5.33DD177 pKa = 7.3DD178 pKa = 7.36DD179 pKa = 6.42DD180 pKa = 6.72DD181 pKa = 6.66YY182 pKa = 12.15YY183 pKa = 11.65DD184 pKa = 6.18DD185 pKa = 5.89DD186 pKa = 6.84DD187 pKa = 6.05NDD189 pKa = 3.73EE190 pKa = 4.39TCQPEE195 pKa = 4.37QEE197 pKa = 4.7DD198 pKa = 3.88EE199 pKa = 4.37HH200 pKa = 8.57NEE202 pKa = 3.83AEE204 pKa = 4.96PIDD207 pKa = 4.25EE208 pKa = 4.32EE209 pKa = 5.18SFTLSEE215 pKa = 4.73DD216 pKa = 4.06VNSSDD221 pKa = 3.37YY222 pKa = 9.29DD223 pKa = 4.07TPVHH227 pKa = 6.7VYY229 pKa = 10.1FDD231 pKa = 4.09DD232 pKa = 5.26RR233 pKa = 11.84EE234 pKa = 4.23DD235 pKa = 3.45VYY237 pKa = 10.92FASEE241 pKa = 4.11CCEE244 pKa = 3.92THH246 pKa = 6.69QLTEE250 pKa = 4.21VNSPHH255 pKa = 6.84DD256 pKa = 4.36TLNMSADD263 pKa = 3.43HH264 pKa = 7.25CEE266 pKa = 3.97MCEE269 pKa = 3.75TDD271 pKa = 3.94YY272 pKa = 11.77SEE274 pKa = 4.44TSQEE278 pKa = 3.79YY279 pKa = 10.1DD280 pKa = 3.43QQCATSKK287 pKa = 10.06QCSHH291 pKa = 6.24KK292 pKa = 9.14TSEE295 pKa = 4.26FGMEE299 pKa = 3.97EE300 pKa = 4.71DD301 pKa = 4.54GSSDD305 pKa = 3.44CSSIEE310 pKa = 3.94TKK312 pKa = 10.48SFKK315 pKa = 9.84TCPDD319 pKa = 2.84GSIPSDD325 pKa = 3.08ICSDD329 pKa = 3.83SSGEE333 pKa = 4.25SQKK336 pKa = 7.85TTQEE340 pKa = 4.13YY341 pKa = 10.78SSDD344 pKa = 3.34EE345 pKa = 4.14HH346 pKa = 5.35THH348 pKa = 5.65WEE350 pKa = 4.15SFEE353 pKa = 4.09EE354 pKa = 4.46DD355 pKa = 3.98DD356 pKa = 5.63EE357 pKa = 5.52IEE359 pKa = 3.99QSSINGSNEE368 pKa = 3.77DD369 pKa = 3.41QTITPTVNSVIEE381 pKa = 3.98DD382 pKa = 3.59HH383 pKa = 7.38FDD385 pKa = 3.31LFDD388 pKa = 4.57RR389 pKa = 11.84ADD391 pKa = 3.29YY392 pKa = 9.85WGNTFAQKK400 pKa = 10.1QRR402 pKa = 11.84YY403 pKa = 8.24ISCFDD408 pKa = 3.58GGDD411 pKa = 3.0IHH413 pKa = 7.65DD414 pKa = 4.39RR415 pKa = 11.84LHH417 pKa = 8.05LEE419 pKa = 4.28EE420 pKa = 4.54VQSEE424 pKa = 4.25AQKK427 pKa = 10.48CAKK430 pKa = 9.72NVYY433 pKa = 9.33KK434 pKa = 10.65FKK436 pKa = 10.37EE437 pKa = 3.89IKK439 pKa = 9.15KK440 pKa = 8.49TIDD443 pKa = 3.29VLEE446 pKa = 4.57TCLDD450 pKa = 3.7APEE453 pKa = 4.38EE454 pKa = 4.34ACEE457 pKa = 3.92DD458 pKa = 3.98TYY460 pKa = 11.79EE461 pKa = 4.21EE462 pKa = 4.98DD463 pKa = 3.81ASQRR467 pKa = 11.84DD468 pKa = 3.82DD469 pKa = 4.21ASTGSCEE476 pKa = 4.58SEE478 pKa = 3.97EE479 pKa = 4.72QAEE482 pKa = 4.18DD483 pKa = 3.65WIKK486 pKa = 11.04EE487 pKa = 4.3SGSSLSGDD495 pKa = 3.22EE496 pKa = 4.28VFVDD500 pKa = 4.15EE501 pKa = 5.23SEE503 pKa = 4.54VFALYY508 pKa = 10.5EE509 pKa = 4.21EE510 pKa = 4.52NCEE513 pKa = 4.0EE514 pKa = 5.41DD515 pKa = 4.07EE516 pKa = 4.19DD517 pKa = 4.45TALDD521 pKa = 3.58GHH523 pKa = 5.38VTEE526 pKa = 5.63IYY528 pKa = 10.46NEE530 pKa = 4.06EE531 pKa = 4.0EE532 pKa = 4.66AEE534 pKa = 4.25VCLPSGNKK542 pKa = 9.69EE543 pKa = 4.22SMSAPCADD551 pKa = 5.01DD552 pKa = 3.64ISVEE556 pKa = 4.14EE557 pKa = 4.34VEE559 pKa = 4.97PYY561 pKa = 9.52WLLVDD566 pKa = 4.08HH567 pKa = 7.09EE568 pKa = 4.76EE569 pKa = 4.14NEE571 pKa = 4.55EE572 pKa = 3.98IVITQLTPHH581 pKa = 6.67EE582 pKa = 4.58ARR584 pKa = 11.84TEE586 pKa = 4.0EE587 pKa = 4.19NKK589 pKa = 10.62ASEE592 pKa = 4.49STDD595 pKa = 3.14SDD597 pKa = 3.28EE598 pKa = 6.31DD599 pKa = 3.41EE600 pKa = 4.36WSDD603 pKa = 3.89EE604 pKa = 4.0ISTEE608 pKa = 3.94LCEE611 pKa = 5.05CEE613 pKa = 4.36YY614 pKa = 10.59CIPPIEE620 pKa = 4.34EE621 pKa = 4.12VPAKK625 pKa = 10.26PLLPRR630 pKa = 11.84MKK632 pKa = 9.86STDD635 pKa = 3.05AGKK638 pKa = 9.43ICVVIDD644 pKa = 3.71LDD646 pKa = 3.74EE647 pKa = 4.47TLVHH651 pKa = 6.68SSFKK655 pKa = 10.96VYY657 pKa = 9.99VLKK660 pKa = 10.64RR661 pKa = 11.84PHH663 pKa = 7.15VDD665 pKa = 2.95EE666 pKa = 4.17FLKK669 pKa = 10.99RR670 pKa = 11.84MGEE673 pKa = 3.96LFEE676 pKa = 5.41CVLFTASLSKK686 pKa = 11.06YY687 pKa = 9.64ADD689 pKa = 3.7PVSDD693 pKa = 6.39LLDD696 pKa = 2.94KK697 pKa = 10.75WGAFRR702 pKa = 11.84SRR704 pKa = 11.84LFRR707 pKa = 11.84EE708 pKa = 3.9SCVFHH713 pKa = 6.89KK714 pKa = 10.9GNYY717 pKa = 9.11VKK719 pKa = 10.65DD720 pKa = 3.95LSRR723 pKa = 11.84LGRR726 pKa = 11.84DD727 pKa = 3.29LNKK730 pKa = 10.25VIIIDD735 pKa = 3.96NSPASYY741 pKa = 10.77VFHH744 pKa = 7.92PDD746 pKa = 2.61NAVPVASWFDD756 pKa = 3.67DD757 pKa = 3.6MSDD760 pKa = 3.58TEE762 pKa = 5.18LLDD765 pKa = 4.78LIPFFEE771 pKa = 4.75RR772 pKa = 11.84LSKK775 pKa = 10.86VDD777 pKa = 5.0DD778 pKa = 4.11IYY780 pKa = 11.59EE781 pKa = 4.03VLQQQRR787 pKa = 11.84TSSS790 pKa = 3.19
MM1 pKa = 7.73DD2 pKa = 5.74HH3 pKa = 6.77SPSIITQVTRR13 pKa = 11.84DD14 pKa = 3.57EE15 pKa = 4.6EE16 pKa = 5.03DD17 pKa = 3.16NATCRR22 pKa = 11.84EE23 pKa = 4.08DD24 pKa = 3.4GANEE28 pKa = 3.7VSTSKK33 pKa = 10.59KK34 pKa = 8.14PRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GLLDD42 pKa = 3.85SLFCCLCRR50 pKa = 11.84KK51 pKa = 9.23EE52 pKa = 5.13SEE54 pKa = 4.28PPPLKK59 pKa = 10.92NNAPLLVEE67 pKa = 4.34EE68 pKa = 5.1NGTLSKK74 pKa = 10.43QCKK77 pKa = 9.54PSDD80 pKa = 3.63QQFEE84 pKa = 4.41SSDD87 pKa = 3.87FEE89 pKa = 5.82PDD91 pKa = 3.17TLAVNHH97 pKa = 7.1DD98 pKa = 3.83GFGQCEE104 pKa = 4.05MTDD107 pKa = 4.4FIPEE111 pKa = 4.35CTDD114 pKa = 3.35HH115 pKa = 8.14LDD117 pKa = 3.65FLQHH121 pKa = 6.31YY122 pKa = 7.21EE123 pKa = 4.09PCDD126 pKa = 3.53PQCEE130 pKa = 4.28YY131 pKa = 11.13FEE133 pKa = 5.97PDD135 pKa = 3.85TSTEE139 pKa = 3.88DD140 pKa = 3.35CKK142 pKa = 10.88QCEE145 pKa = 4.26RR146 pKa = 11.84TGFTPNVSDD155 pKa = 5.82SEE157 pKa = 4.57DD158 pKa = 3.65LLDD161 pKa = 5.15CGTEE165 pKa = 3.62HH166 pKa = 7.47FEE168 pKa = 5.02YY169 pKa = 10.83DD170 pKa = 3.31EE171 pKa = 4.61CTNDD175 pKa = 5.61DD176 pKa = 5.33DD177 pKa = 7.3DD178 pKa = 7.36DD179 pKa = 6.42DD180 pKa = 6.72DD181 pKa = 6.66YY182 pKa = 12.15YY183 pKa = 11.65DD184 pKa = 6.18DD185 pKa = 5.89DD186 pKa = 6.84DD187 pKa = 6.05NDD189 pKa = 3.73EE190 pKa = 4.39TCQPEE195 pKa = 4.37QEE197 pKa = 4.7DD198 pKa = 3.88EE199 pKa = 4.37HH200 pKa = 8.57NEE202 pKa = 3.83AEE204 pKa = 4.96PIDD207 pKa = 4.25EE208 pKa = 4.32EE209 pKa = 5.18SFTLSEE215 pKa = 4.73DD216 pKa = 4.06VNSSDD221 pKa = 3.37YY222 pKa = 9.29DD223 pKa = 4.07TPVHH227 pKa = 6.7VYY229 pKa = 10.1FDD231 pKa = 4.09DD232 pKa = 5.26RR233 pKa = 11.84EE234 pKa = 4.23DD235 pKa = 3.45VYY237 pKa = 10.92FASEE241 pKa = 4.11CCEE244 pKa = 3.92THH246 pKa = 6.69QLTEE250 pKa = 4.21VNSPHH255 pKa = 6.84DD256 pKa = 4.36TLNMSADD263 pKa = 3.43HH264 pKa = 7.25CEE266 pKa = 3.97MCEE269 pKa = 3.75TDD271 pKa = 3.94YY272 pKa = 11.77SEE274 pKa = 4.44TSQEE278 pKa = 3.79YY279 pKa = 10.1DD280 pKa = 3.43QQCATSKK287 pKa = 10.06QCSHH291 pKa = 6.24KK292 pKa = 9.14TSEE295 pKa = 4.26FGMEE299 pKa = 3.97EE300 pKa = 4.71DD301 pKa = 4.54GSSDD305 pKa = 3.44CSSIEE310 pKa = 3.94TKK312 pKa = 10.48SFKK315 pKa = 9.84TCPDD319 pKa = 2.84GSIPSDD325 pKa = 3.08ICSDD329 pKa = 3.83SSGEE333 pKa = 4.25SQKK336 pKa = 7.85TTQEE340 pKa = 4.13YY341 pKa = 10.78SSDD344 pKa = 3.34EE345 pKa = 4.14HH346 pKa = 5.35THH348 pKa = 5.65WEE350 pKa = 4.15SFEE353 pKa = 4.09EE354 pKa = 4.46DD355 pKa = 3.98DD356 pKa = 5.63EE357 pKa = 5.52IEE359 pKa = 3.99QSSINGSNEE368 pKa = 3.77DD369 pKa = 3.41QTITPTVNSVIEE381 pKa = 3.98DD382 pKa = 3.59HH383 pKa = 7.38FDD385 pKa = 3.31LFDD388 pKa = 4.57RR389 pKa = 11.84ADD391 pKa = 3.29YY392 pKa = 9.85WGNTFAQKK400 pKa = 10.1QRR402 pKa = 11.84YY403 pKa = 8.24ISCFDD408 pKa = 3.58GGDD411 pKa = 3.0IHH413 pKa = 7.65DD414 pKa = 4.39RR415 pKa = 11.84LHH417 pKa = 8.05LEE419 pKa = 4.28EE420 pKa = 4.54VQSEE424 pKa = 4.25AQKK427 pKa = 10.48CAKK430 pKa = 9.72NVYY433 pKa = 9.33KK434 pKa = 10.65FKK436 pKa = 10.37EE437 pKa = 3.89IKK439 pKa = 9.15KK440 pKa = 8.49TIDD443 pKa = 3.29VLEE446 pKa = 4.57TCLDD450 pKa = 3.7APEE453 pKa = 4.38EE454 pKa = 4.34ACEE457 pKa = 3.92DD458 pKa = 3.98TYY460 pKa = 11.79EE461 pKa = 4.21EE462 pKa = 4.98DD463 pKa = 3.81ASQRR467 pKa = 11.84DD468 pKa = 3.82DD469 pKa = 4.21ASTGSCEE476 pKa = 4.58SEE478 pKa = 3.97EE479 pKa = 4.72QAEE482 pKa = 4.18DD483 pKa = 3.65WIKK486 pKa = 11.04EE487 pKa = 4.3SGSSLSGDD495 pKa = 3.22EE496 pKa = 4.28VFVDD500 pKa = 4.15EE501 pKa = 5.23SEE503 pKa = 4.54VFALYY508 pKa = 10.5EE509 pKa = 4.21EE510 pKa = 4.52NCEE513 pKa = 4.0EE514 pKa = 5.41DD515 pKa = 4.07EE516 pKa = 4.19DD517 pKa = 4.45TALDD521 pKa = 3.58GHH523 pKa = 5.38VTEE526 pKa = 5.63IYY528 pKa = 10.46NEE530 pKa = 4.06EE531 pKa = 4.0EE532 pKa = 4.66AEE534 pKa = 4.25VCLPSGNKK542 pKa = 9.69EE543 pKa = 4.22SMSAPCADD551 pKa = 5.01DD552 pKa = 3.64ISVEE556 pKa = 4.14EE557 pKa = 4.34VEE559 pKa = 4.97PYY561 pKa = 9.52WLLVDD566 pKa = 4.08HH567 pKa = 7.09EE568 pKa = 4.76EE569 pKa = 4.14NEE571 pKa = 4.55EE572 pKa = 3.98IVITQLTPHH581 pKa = 6.67EE582 pKa = 4.58ARR584 pKa = 11.84TEE586 pKa = 4.0EE587 pKa = 4.19NKK589 pKa = 10.62ASEE592 pKa = 4.49STDD595 pKa = 3.14SDD597 pKa = 3.28EE598 pKa = 6.31DD599 pKa = 3.41EE600 pKa = 4.36WSDD603 pKa = 3.89EE604 pKa = 4.0ISTEE608 pKa = 3.94LCEE611 pKa = 5.05CEE613 pKa = 4.36YY614 pKa = 10.59CIPPIEE620 pKa = 4.34EE621 pKa = 4.12VPAKK625 pKa = 10.26PLLPRR630 pKa = 11.84MKK632 pKa = 9.86STDD635 pKa = 3.05AGKK638 pKa = 9.43ICVVIDD644 pKa = 3.71LDD646 pKa = 3.74EE647 pKa = 4.47TLVHH651 pKa = 6.68SSFKK655 pKa = 10.96VYY657 pKa = 9.99VLKK660 pKa = 10.64RR661 pKa = 11.84PHH663 pKa = 7.15VDD665 pKa = 2.95EE666 pKa = 4.17FLKK669 pKa = 10.99RR670 pKa = 11.84MGEE673 pKa = 3.96LFEE676 pKa = 5.41CVLFTASLSKK686 pKa = 11.06YY687 pKa = 9.64ADD689 pKa = 3.7PVSDD693 pKa = 6.39LLDD696 pKa = 2.94KK697 pKa = 10.75WGAFRR702 pKa = 11.84SRR704 pKa = 11.84LFRR707 pKa = 11.84EE708 pKa = 3.9SCVFHH713 pKa = 6.89KK714 pKa = 10.9GNYY717 pKa = 9.11VKK719 pKa = 10.65DD720 pKa = 3.95LSRR723 pKa = 11.84LGRR726 pKa = 11.84DD727 pKa = 3.29LNKK730 pKa = 10.25VIIIDD735 pKa = 3.96NSPASYY741 pKa = 10.77VFHH744 pKa = 7.92PDD746 pKa = 2.61NAVPVASWFDD756 pKa = 3.67DD757 pKa = 3.6MSDD760 pKa = 3.58TEE762 pKa = 5.18LLDD765 pKa = 4.78LIPFFEE771 pKa = 4.75RR772 pKa = 11.84LSKK775 pKa = 10.86VDD777 pKa = 5.0DD778 pKa = 4.11IYY780 pKa = 11.59EE781 pKa = 4.03VLQQQRR787 pKa = 11.84TSSS790 pKa = 3.19
Molecular weight: 89.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J5D9M3|A0A5J5D9M3_9PERO TAFH domain-containing protein OS=Etheostoma spectabile OX=54343 GN=FQN60_001790 PE=3 SV=1
MM1 pKa = 8.12RR2 pKa = 11.84ITSVLRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84AWWHH14 pKa = 4.69GRR16 pKa = 11.84ATRR19 pKa = 11.84HH20 pKa = 6.2HH21 pKa = 6.98LFQRR25 pKa = 11.84GTT27 pKa = 3.29
MM1 pKa = 8.12RR2 pKa = 11.84ITSVLRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84AWWHH14 pKa = 4.69GRR16 pKa = 11.84ATRR19 pKa = 11.84HH20 pKa = 6.2HH21 pKa = 6.98LFQRR25 pKa = 11.84GTT27 pKa = 3.29
Molecular weight: 3.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8925540 |
9 |
17660 |
485.8 |
54.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.639 ± 0.014 | 2.274 ± 0.015 |
5.078 ± 0.014 | 6.768 ± 0.027 |
3.459 ± 0.014 | 6.46 ± 0.021 |
2.723 ± 0.011 | 4.18 ± 0.015 |
5.463 ± 0.022 | 9.543 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.378 ± 0.008 | 3.764 ± 0.01 |
5.911 ± 0.021 | 4.804 ± 0.021 |
5.751 ± 0.014 | 8.934 ± 0.025 |
5.741 ± 0.017 | 6.335 ± 0.02 |
1.183 ± 0.007 | 2.593 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
