
Vibrio phage phiVa1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
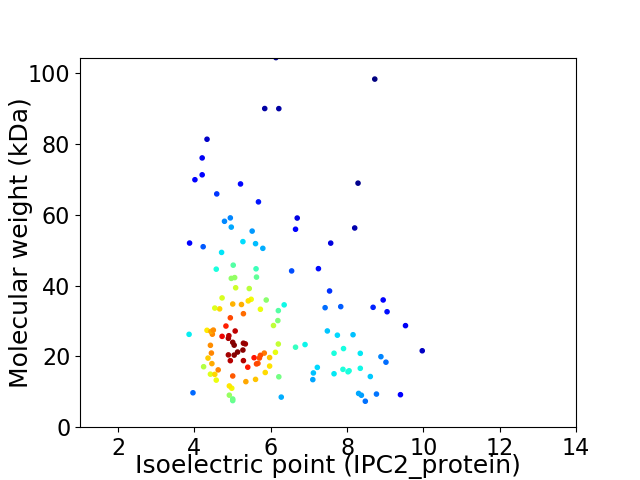
Virtual 2D-PAGE plot for 135 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A410T6Z5|A0A410T6Z5_9CAUD Homing endonuclease OS=Vibrio phage phiVa1 OX=2508852 GN=ValB1MD_319 PE=4 SV=1
MM1 pKa = 7.69ARR3 pKa = 11.84DD4 pKa = 3.88LKK6 pKa = 11.31NIFDD10 pKa = 5.01SITPEE15 pKa = 4.17NIKK18 pKa = 10.35DD19 pKa = 3.52VPLIEE24 pKa = 4.72DD25 pKa = 3.37AMSIFIEE32 pKa = 4.26SLEE35 pKa = 4.09EE36 pKa = 3.82LSKK39 pKa = 11.26EE40 pKa = 4.26SIDD43 pKa = 3.84VKK45 pKa = 10.73NVYY48 pKa = 10.35SNEE51 pKa = 4.08IIRR54 pKa = 11.84EE55 pKa = 4.03EE56 pKa = 4.35LIKK59 pKa = 10.31IYY61 pKa = 10.69LDD63 pKa = 3.2DD64 pKa = 4.78LYY66 pKa = 11.29RR67 pKa = 11.84VFKK70 pKa = 10.75NIQLNQKK77 pKa = 9.92IIDD80 pKa = 4.84RR81 pKa = 11.84IDD83 pKa = 3.5SLNGIYY89 pKa = 10.66GNNDD93 pKa = 2.58QYY95 pKa = 11.7FNKK98 pKa = 10.42DD99 pKa = 3.31AILNMASYY107 pKa = 11.29LNDD110 pKa = 3.25EE111 pKa = 4.51HH112 pKa = 8.23FMTIKK117 pKa = 10.5AFKK120 pKa = 9.46EE121 pKa = 4.2SKK123 pKa = 9.08GTEE126 pKa = 3.63KK127 pKa = 10.59AIEE130 pKa = 4.04YY131 pKa = 8.14MYY133 pKa = 11.23SLISVFVSPSDD144 pKa = 3.44NGEE147 pKa = 3.98SFQLIEE153 pKa = 4.7NDD155 pKa = 3.26VFDD158 pKa = 3.71YY159 pKa = 9.96TIRR162 pKa = 11.84GEE164 pKa = 4.18LPEE167 pKa = 4.56EE168 pKa = 4.16FYY170 pKa = 11.48NYY172 pKa = 9.3IVAPLAHH179 pKa = 7.23PLGFTYY185 pKa = 9.72TYY187 pKa = 9.73EE188 pKa = 4.13QYY190 pKa = 11.33LSLVLEE196 pKa = 4.63DD197 pKa = 5.26FFPGEE202 pKa = 4.2NISYY206 pKa = 9.87DD207 pKa = 3.35INALEE212 pKa = 4.23VRR214 pKa = 11.84CLFPDD219 pKa = 3.99GTTKK223 pKa = 9.4VTDD226 pKa = 4.04FKK228 pKa = 11.48YY229 pKa = 11.04YY230 pKa = 10.35YY231 pKa = 9.87DD232 pKa = 3.77EE233 pKa = 4.61NGNIIKK239 pKa = 10.24DD240 pKa = 3.79EE241 pKa = 4.4NGTPVQRR248 pKa = 11.84VVVDD252 pKa = 3.25VTVINDD258 pKa = 2.97VGEE261 pKa = 4.06RR262 pKa = 11.84RR263 pKa = 11.84KK264 pKa = 10.58KK265 pKa = 10.33ILFEE269 pKa = 5.28DD270 pKa = 4.01GTYY273 pKa = 11.02LLQVSLINGRR283 pKa = 11.84TNVYY287 pKa = 10.37FMDD290 pKa = 4.52EE291 pKa = 3.73NDD293 pKa = 3.26EE294 pKa = 4.51TIKK297 pKa = 11.03HH298 pKa = 6.5FDD300 pKa = 3.69QQCSIYY306 pKa = 10.5FEE308 pKa = 4.99YY309 pKa = 10.56EE310 pKa = 3.65FNYY313 pKa = 9.15DD314 pKa = 3.26TTLGEE319 pKa = 4.31EE320 pKa = 4.69FSSEE324 pKa = 4.12DD325 pKa = 3.45TKK327 pKa = 11.07STSDD331 pKa = 3.11YY332 pKa = 10.53FARR335 pKa = 11.84LNSDD339 pKa = 3.01AAAIYY344 pKa = 10.05IGLEE348 pKa = 4.14DD349 pKa = 6.12DD350 pKa = 4.13PTLDD354 pKa = 5.27DD355 pKa = 3.9IGEE358 pKa = 4.15FFIGNQLDD366 pKa = 3.8LFDD369 pKa = 5.35IGFNLIRR376 pKa = 11.84DD377 pKa = 3.59RR378 pKa = 11.84DD379 pKa = 4.08GNNLIPSGMQGEE391 pKa = 4.14EE392 pKa = 4.03HH393 pKa = 6.31GRR395 pKa = 11.84KK396 pKa = 8.0EE397 pKa = 4.6HH398 pKa = 6.14YY399 pKa = 9.88FSPSEE404 pKa = 3.99TADD407 pKa = 3.59LKK409 pKa = 11.44VLLGKK414 pKa = 9.02YY415 pKa = 9.34EE416 pKa = 4.13KK417 pKa = 10.88DD418 pKa = 2.76VDD420 pKa = 3.11IFYY423 pKa = 10.78NGEE426 pKa = 3.94YY427 pKa = 10.73LEE429 pKa = 4.46TQHH432 pKa = 7.41IFYY435 pKa = 10.2TDD437 pKa = 4.1IINPSYY443 pKa = 10.76QVVEE447 pKa = 4.26RR448 pKa = 11.84VNSITKK454 pKa = 9.84YY455 pKa = 9.48QEE457 pKa = 3.75QNIGNEE463 pKa = 4.08DD464 pKa = 3.9YY465 pKa = 10.76LAHH468 pKa = 6.12EE469 pKa = 4.48HH470 pKa = 6.14MIEE473 pKa = 4.31NIEE476 pKa = 4.37SLATYY481 pKa = 9.81NQITLEE487 pKa = 3.89PAYY490 pKa = 10.82VGTHH494 pKa = 5.49NFIDD498 pKa = 3.8ANGVNRR504 pKa = 11.84GAFPGYY510 pKa = 9.57MIIGGNYY517 pKa = 10.25DD518 pKa = 3.39FDD520 pKa = 4.81RR521 pKa = 11.84SQTEE525 pKa = 4.02VEE527 pKa = 4.16QVIGYY532 pKa = 10.07DD533 pKa = 3.3PFTNDD538 pKa = 2.42GTFIGGYY545 pKa = 9.87SDD547 pKa = 4.35DD548 pKa = 3.49QDD550 pKa = 4.09YY551 pKa = 11.95NEE553 pKa = 5.48DD554 pKa = 3.35LSQTVNPNEE563 pKa = 4.07SIIYY567 pKa = 10.51FEE569 pKa = 4.7TNVDD573 pKa = 3.41ADD575 pKa = 4.04SVNLQVEE582 pKa = 4.75TYY584 pKa = 10.89DD585 pKa = 3.85VFDD588 pKa = 4.46CGVYY592 pKa = 9.93RR593 pKa = 11.84DD594 pKa = 4.09GVLLEE599 pKa = 5.17DD600 pKa = 4.22NNISALSS607 pKa = 3.5
MM1 pKa = 7.69ARR3 pKa = 11.84DD4 pKa = 3.88LKK6 pKa = 11.31NIFDD10 pKa = 5.01SITPEE15 pKa = 4.17NIKK18 pKa = 10.35DD19 pKa = 3.52VPLIEE24 pKa = 4.72DD25 pKa = 3.37AMSIFIEE32 pKa = 4.26SLEE35 pKa = 4.09EE36 pKa = 3.82LSKK39 pKa = 11.26EE40 pKa = 4.26SIDD43 pKa = 3.84VKK45 pKa = 10.73NVYY48 pKa = 10.35SNEE51 pKa = 4.08IIRR54 pKa = 11.84EE55 pKa = 4.03EE56 pKa = 4.35LIKK59 pKa = 10.31IYY61 pKa = 10.69LDD63 pKa = 3.2DD64 pKa = 4.78LYY66 pKa = 11.29RR67 pKa = 11.84VFKK70 pKa = 10.75NIQLNQKK77 pKa = 9.92IIDD80 pKa = 4.84RR81 pKa = 11.84IDD83 pKa = 3.5SLNGIYY89 pKa = 10.66GNNDD93 pKa = 2.58QYY95 pKa = 11.7FNKK98 pKa = 10.42DD99 pKa = 3.31AILNMASYY107 pKa = 11.29LNDD110 pKa = 3.25EE111 pKa = 4.51HH112 pKa = 8.23FMTIKK117 pKa = 10.5AFKK120 pKa = 9.46EE121 pKa = 4.2SKK123 pKa = 9.08GTEE126 pKa = 3.63KK127 pKa = 10.59AIEE130 pKa = 4.04YY131 pKa = 8.14MYY133 pKa = 11.23SLISVFVSPSDD144 pKa = 3.44NGEE147 pKa = 3.98SFQLIEE153 pKa = 4.7NDD155 pKa = 3.26VFDD158 pKa = 3.71YY159 pKa = 9.96TIRR162 pKa = 11.84GEE164 pKa = 4.18LPEE167 pKa = 4.56EE168 pKa = 4.16FYY170 pKa = 11.48NYY172 pKa = 9.3IVAPLAHH179 pKa = 7.23PLGFTYY185 pKa = 9.72TYY187 pKa = 9.73EE188 pKa = 4.13QYY190 pKa = 11.33LSLVLEE196 pKa = 4.63DD197 pKa = 5.26FFPGEE202 pKa = 4.2NISYY206 pKa = 9.87DD207 pKa = 3.35INALEE212 pKa = 4.23VRR214 pKa = 11.84CLFPDD219 pKa = 3.99GTTKK223 pKa = 9.4VTDD226 pKa = 4.04FKK228 pKa = 11.48YY229 pKa = 11.04YY230 pKa = 10.35YY231 pKa = 9.87DD232 pKa = 3.77EE233 pKa = 4.61NGNIIKK239 pKa = 10.24DD240 pKa = 3.79EE241 pKa = 4.4NGTPVQRR248 pKa = 11.84VVVDD252 pKa = 3.25VTVINDD258 pKa = 2.97VGEE261 pKa = 4.06RR262 pKa = 11.84RR263 pKa = 11.84KK264 pKa = 10.58KK265 pKa = 10.33ILFEE269 pKa = 5.28DD270 pKa = 4.01GTYY273 pKa = 11.02LLQVSLINGRR283 pKa = 11.84TNVYY287 pKa = 10.37FMDD290 pKa = 4.52EE291 pKa = 3.73NDD293 pKa = 3.26EE294 pKa = 4.51TIKK297 pKa = 11.03HH298 pKa = 6.5FDD300 pKa = 3.69QQCSIYY306 pKa = 10.5FEE308 pKa = 4.99YY309 pKa = 10.56EE310 pKa = 3.65FNYY313 pKa = 9.15DD314 pKa = 3.26TTLGEE319 pKa = 4.31EE320 pKa = 4.69FSSEE324 pKa = 4.12DD325 pKa = 3.45TKK327 pKa = 11.07STSDD331 pKa = 3.11YY332 pKa = 10.53FARR335 pKa = 11.84LNSDD339 pKa = 3.01AAAIYY344 pKa = 10.05IGLEE348 pKa = 4.14DD349 pKa = 6.12DD350 pKa = 4.13PTLDD354 pKa = 5.27DD355 pKa = 3.9IGEE358 pKa = 4.15FFIGNQLDD366 pKa = 3.8LFDD369 pKa = 5.35IGFNLIRR376 pKa = 11.84DD377 pKa = 3.59RR378 pKa = 11.84DD379 pKa = 4.08GNNLIPSGMQGEE391 pKa = 4.14EE392 pKa = 4.03HH393 pKa = 6.31GRR395 pKa = 11.84KK396 pKa = 8.0EE397 pKa = 4.6HH398 pKa = 6.14YY399 pKa = 9.88FSPSEE404 pKa = 3.99TADD407 pKa = 3.59LKK409 pKa = 11.44VLLGKK414 pKa = 9.02YY415 pKa = 9.34EE416 pKa = 4.13KK417 pKa = 10.88DD418 pKa = 2.76VDD420 pKa = 3.11IFYY423 pKa = 10.78NGEE426 pKa = 3.94YY427 pKa = 10.73LEE429 pKa = 4.46TQHH432 pKa = 7.41IFYY435 pKa = 10.2TDD437 pKa = 4.1IINPSYY443 pKa = 10.76QVVEE447 pKa = 4.26RR448 pKa = 11.84VNSITKK454 pKa = 9.84YY455 pKa = 9.48QEE457 pKa = 3.75QNIGNEE463 pKa = 4.08DD464 pKa = 3.9YY465 pKa = 10.76LAHH468 pKa = 6.12EE469 pKa = 4.48HH470 pKa = 6.14MIEE473 pKa = 4.31NIEE476 pKa = 4.37SLATYY481 pKa = 9.81NQITLEE487 pKa = 3.89PAYY490 pKa = 10.82VGTHH494 pKa = 5.49NFIDD498 pKa = 3.8ANGVNRR504 pKa = 11.84GAFPGYY510 pKa = 9.57MIIGGNYY517 pKa = 10.25DD518 pKa = 3.39FDD520 pKa = 4.81RR521 pKa = 11.84SQTEE525 pKa = 4.02VEE527 pKa = 4.16QVIGYY532 pKa = 10.07DD533 pKa = 3.3PFTNDD538 pKa = 2.42GTFIGGYY545 pKa = 9.87SDD547 pKa = 4.35DD548 pKa = 3.49QDD550 pKa = 4.09YY551 pKa = 11.95NEE553 pKa = 5.48DD554 pKa = 3.35LSQTVNPNEE563 pKa = 4.07SIIYY567 pKa = 10.51FEE569 pKa = 4.7TNVDD573 pKa = 3.41ADD575 pKa = 4.04SVNLQVEE582 pKa = 4.75TYY584 pKa = 10.89DD585 pKa = 3.85VFDD588 pKa = 4.46CGVYY592 pKa = 9.93RR593 pKa = 11.84DD594 pKa = 4.09GVLLEE599 pKa = 5.17DD600 pKa = 4.22NNISALSS607 pKa = 3.5
Molecular weight: 69.87 kDa
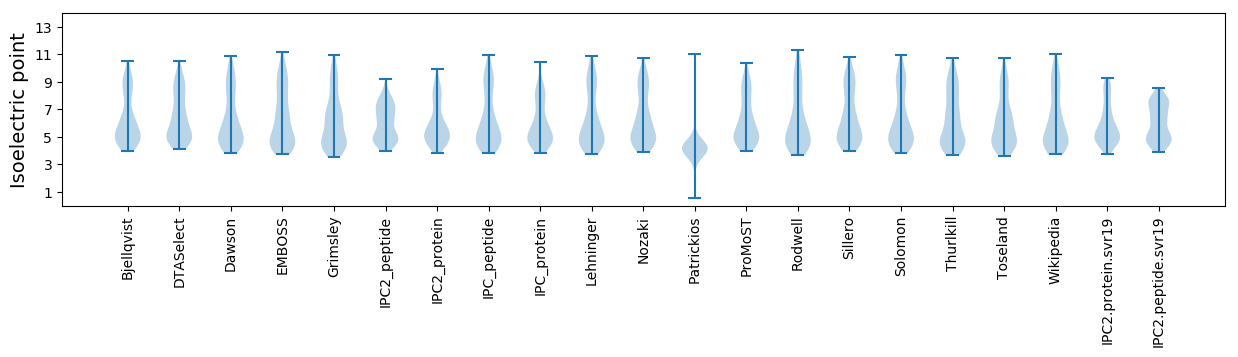
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A410T6I8|A0A410T6I8_9CAUD DUF2493 domain-containing protein OS=Vibrio phage phiVa1 OX=2508852 GN=ValB1MD_021 PE=4 SV=1
MM1 pKa = 7.2ARR3 pKa = 11.84KK4 pKa = 8.62VQKK7 pKa = 9.98EE8 pKa = 4.04LTPAEE13 pKa = 4.07SVKK16 pKa = 10.29IVKK19 pKa = 10.39KK20 pKa = 10.36LVKK23 pKa = 9.99DD24 pKa = 3.49KK25 pKa = 11.34KK26 pKa = 10.5RR27 pKa = 11.84INKK30 pKa = 9.39ADD32 pKa = 3.68WVPGNIIYY40 pKa = 8.97TYY42 pKa = 11.04YY43 pKa = 10.33LAKK46 pKa = 10.9DD47 pKa = 3.26MTQVYY52 pKa = 10.61DD53 pKa = 3.63RR54 pKa = 11.84TPLVLILRR62 pKa = 11.84RR63 pKa = 11.84NKK65 pKa = 8.79THH67 pKa = 6.17TLGLNFHH74 pKa = 7.6WIPFKK79 pKa = 10.83MRR81 pKa = 11.84DD82 pKa = 3.16NLARR86 pKa = 11.84YY87 pKa = 8.66ILKK90 pKa = 10.41LNKK93 pKa = 9.8KK94 pKa = 9.83NIEE97 pKa = 3.83QGKK100 pKa = 8.26PLEE103 pKa = 3.98FSYY106 pKa = 10.95RR107 pKa = 11.84QLKK110 pKa = 9.83PILKK114 pKa = 9.1NYY116 pKa = 9.81GYY118 pKa = 10.63APCIRR123 pKa = 11.84LYY125 pKa = 10.18INHH128 pKa = 7.4RR129 pKa = 11.84FRR131 pKa = 11.84SSGTVIPPDD140 pKa = 3.21RR141 pKa = 11.84LVEE144 pKa = 4.09VARR147 pKa = 11.84LRR149 pKa = 11.84TEE151 pKa = 3.82TFTKK155 pKa = 10.46GKK157 pKa = 8.3YY158 pKa = 7.31TAEE161 pKa = 3.75QLYY164 pKa = 10.82AKK166 pKa = 9.89AVKK169 pKa = 10.18DD170 pKa = 3.44GRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84AKK176 pKa = 9.76ARR178 pKa = 11.84KK179 pKa = 8.92RR180 pKa = 11.84KK181 pKa = 9.37KK182 pKa = 10.26
MM1 pKa = 7.2ARR3 pKa = 11.84KK4 pKa = 8.62VQKK7 pKa = 9.98EE8 pKa = 4.04LTPAEE13 pKa = 4.07SVKK16 pKa = 10.29IVKK19 pKa = 10.39KK20 pKa = 10.36LVKK23 pKa = 9.99DD24 pKa = 3.49KK25 pKa = 11.34KK26 pKa = 10.5RR27 pKa = 11.84INKK30 pKa = 9.39ADD32 pKa = 3.68WVPGNIIYY40 pKa = 8.97TYY42 pKa = 11.04YY43 pKa = 10.33LAKK46 pKa = 10.9DD47 pKa = 3.26MTQVYY52 pKa = 10.61DD53 pKa = 3.63RR54 pKa = 11.84TPLVLILRR62 pKa = 11.84RR63 pKa = 11.84NKK65 pKa = 8.79THH67 pKa = 6.17TLGLNFHH74 pKa = 7.6WIPFKK79 pKa = 10.83MRR81 pKa = 11.84DD82 pKa = 3.16NLARR86 pKa = 11.84YY87 pKa = 8.66ILKK90 pKa = 10.41LNKK93 pKa = 9.8KK94 pKa = 9.83NIEE97 pKa = 3.83QGKK100 pKa = 8.26PLEE103 pKa = 3.98FSYY106 pKa = 10.95RR107 pKa = 11.84QLKK110 pKa = 9.83PILKK114 pKa = 9.1NYY116 pKa = 9.81GYY118 pKa = 10.63APCIRR123 pKa = 11.84LYY125 pKa = 10.18INHH128 pKa = 7.4RR129 pKa = 11.84FRR131 pKa = 11.84SSGTVIPPDD140 pKa = 3.21RR141 pKa = 11.84LVEE144 pKa = 4.09VARR147 pKa = 11.84LRR149 pKa = 11.84TEE151 pKa = 3.82TFTKK155 pKa = 10.46GKK157 pKa = 8.3YY158 pKa = 7.31TAEE161 pKa = 3.75QLYY164 pKa = 10.82AKK166 pKa = 9.89AVKK169 pKa = 10.18DD170 pKa = 3.44GRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84AKK176 pKa = 9.76ARR178 pKa = 11.84KK179 pKa = 8.92RR180 pKa = 11.84KK181 pKa = 9.37KK182 pKa = 10.26
Molecular weight: 21.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
37587 |
61 |
896 |
278.4 |
31.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.972 ± 0.253 | 1.019 ± 0.063 |
6.837 ± 0.157 | 7.665 ± 0.201 |
4.781 ± 0.146 | 5.733 ± 0.17 |
1.916 ± 0.109 | 7.284 ± 0.18 |
8.343 ± 0.266 | 7.926 ± 0.168 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.631 ± 0.109 | 6.377 ± 0.166 |
3.11 ± 0.114 | 3.693 ± 0.1 |
3.852 ± 0.129 | 6.901 ± 0.209 |
5.47 ± 0.183 | 6.023 ± 0.138 |
1.014 ± 0.062 | 4.451 ± 0.164 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
