
Rhodobacteraceae bacterium 9Alg 56
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; unclassified Rhodobacteraceae
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
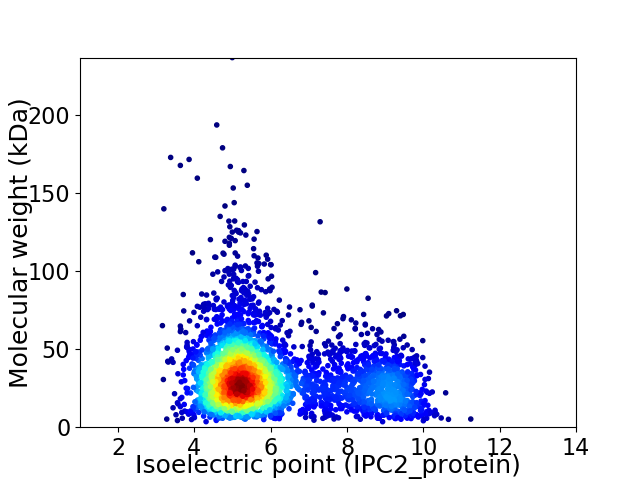
Virtual 2D-PAGE plot for 3843 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P1SXS7|A0A6P1SXS7_9RHOB SCP domain-containing protein OS=Rhodobacteraceae bacterium 9Alg 56 OX=2683284 GN=GO499_09925 PE=4 SV=1
MM1 pKa = 7.27TEE3 pKa = 3.11IRR5 pKa = 11.84NFIFGNSLINHH16 pKa = 5.86VSGSHH21 pKa = 4.44QTTVPYY27 pKa = 10.08WLDD30 pKa = 3.32ALADD34 pKa = 4.16TAGHH38 pKa = 6.3SYY40 pKa = 11.01SVDD43 pKa = 3.24GQFGNLLSHH52 pKa = 7.28LGTNPIPNWWFGDD65 pKa = 3.78GVEE68 pKa = 4.68GSWNSSFANSDD79 pKa = 3.45YY80 pKa = 9.76TNVMITTANYY90 pKa = 9.5IQYY93 pKa = 9.93QGPDD97 pKa = 2.91EE98 pKa = 4.07QYY100 pKa = 10.28YY101 pKa = 10.51GNSHH105 pKa = 5.69TPISATLEE113 pKa = 4.1LVDD116 pKa = 3.86MVHH119 pKa = 6.82ARR121 pKa = 11.84EE122 pKa = 4.08PNAVVDD128 pKa = 5.43IYY130 pKa = 11.61VGWPDD135 pKa = 3.38MAGVFPNFPPSDD147 pKa = 3.85AQFDD151 pKa = 4.3TYY153 pKa = 11.61NQLTMTTYY161 pKa = 9.88MEE163 pKa = 4.61WYY165 pKa = 9.7DD166 pKa = 3.69EE167 pKa = 4.39YY168 pKa = 11.73YY169 pKa = 11.02AALQAARR176 pKa = 11.84PGVTMNLMPVGPAMAHH192 pKa = 6.89LFTTVLSDD200 pKa = 3.16VSAADD205 pKa = 4.05LYY207 pKa = 11.29EE208 pKa = 5.15DD209 pKa = 4.01NAPHH213 pKa = 5.76GTEE216 pKa = 3.66TLYY219 pKa = 11.13FLASLATYY227 pKa = 10.82AFMYY231 pKa = 10.57DD232 pKa = 4.04EE233 pKa = 5.55LPPADD238 pKa = 3.95FQIPNNILPSVRR250 pKa = 11.84AAYY253 pKa = 10.6SDD255 pKa = 3.4ILTEE259 pKa = 4.21LADD262 pKa = 3.79FTGIDD267 pKa = 3.53PAEE270 pKa = 4.17VEE272 pKa = 4.49GGSDD276 pKa = 3.81GDD278 pKa = 3.93SSGGGVDD285 pKa = 4.17VEE287 pKa = 4.57PEE289 pKa = 4.23VNVVDD294 pKa = 5.12GDD296 pKa = 3.86DD297 pKa = 3.56RR298 pKa = 11.84NNNIVGTDD306 pKa = 3.28GMDD309 pKa = 4.3AISGLGGNDD318 pKa = 3.56TIQAGDD324 pKa = 3.7GNDD327 pKa = 4.02TIDD330 pKa = 4.01AGAGNDD336 pKa = 4.78SVWGDD341 pKa = 3.39QGADD345 pKa = 3.62NIHH348 pKa = 6.54GGDD351 pKa = 3.58GSDD354 pKa = 3.55FLSGGRR360 pKa = 11.84GNDD363 pKa = 3.18VVEE366 pKa = 4.77GNSGDD371 pKa = 3.42DD372 pKa = 4.01WINGNYY378 pKa = 10.28GNDD381 pKa = 3.15ILNGGGGNDD390 pKa = 4.35TINAGAGRR398 pKa = 11.84DD399 pKa = 3.63TIYY402 pKa = 10.97AGNGDD407 pKa = 3.57DD408 pKa = 5.05RR409 pKa = 11.84VRR411 pKa = 11.84GGNGRR416 pKa = 11.84DD417 pKa = 3.38TVYY420 pKa = 10.88LGAGHH425 pKa = 7.51DD426 pKa = 3.48RR427 pKa = 11.84FTDD430 pKa = 3.33NGQTGEE436 pKa = 4.19NAVDD440 pKa = 3.49TVYY443 pKa = 11.21GGDD446 pKa = 3.77GDD448 pKa = 4.67DD449 pKa = 5.12AIHH452 pKa = 7.0GGGGDD457 pKa = 3.4DD458 pKa = 3.31RR459 pKa = 11.84FYY461 pKa = 11.74GEE463 pKa = 4.84GGNDD467 pKa = 3.67RR468 pKa = 11.84IFGGSGNDD476 pKa = 3.59QIRR479 pKa = 11.84GGAGDD484 pKa = 4.25DD485 pKa = 3.71SLHH488 pKa = 5.84GRR490 pKa = 11.84EE491 pKa = 4.23GADD494 pKa = 3.62TIFGGNGHH502 pKa = 6.8DD503 pKa = 4.82FINAGSGNDD512 pKa = 3.46TVEE515 pKa = 4.4GGNGRR520 pKa = 11.84DD521 pKa = 3.22RR522 pKa = 11.84VRR524 pKa = 11.84LGGGDD529 pKa = 4.09DD530 pKa = 3.48RR531 pKa = 11.84FVDD534 pKa = 3.43NDD536 pKa = 3.19QAGFQGSDD544 pKa = 3.29TVSGGAGNDD553 pKa = 3.77TILGGGGNDD562 pKa = 3.13RR563 pKa = 11.84LFGGSGADD571 pKa = 3.66TINGGLEE578 pKa = 3.78NDD580 pKa = 4.67HH581 pKa = 6.88ISGGEE586 pKa = 4.03GADD589 pKa = 3.55TLNGGWGDD597 pKa = 3.83DD598 pKa = 4.56FINGAQGADD607 pKa = 3.38IINAGGGDD615 pKa = 3.43DD616 pKa = 4.38HH617 pKa = 8.97VFGGLGADD625 pKa = 3.71VVDD628 pKa = 5.45LGDD631 pKa = 4.52GNDD634 pKa = 3.52IFQDD638 pKa = 3.59SAEE641 pKa = 4.18QGASGADD648 pKa = 3.36TVYY651 pKa = 11.05GGAGNDD657 pKa = 3.72IINGGGGQDD666 pKa = 3.67SLHH669 pKa = 6.67GGDD672 pKa = 4.65GDD674 pKa = 4.17DD675 pKa = 3.64EE676 pKa = 4.58VRR678 pKa = 11.84GGVGNDD684 pKa = 4.56DD685 pKa = 3.46ITGGAGNDD693 pKa = 3.73VLHH696 pKa = 6.92GGAGDD701 pKa = 3.96DD702 pKa = 4.77SIDD705 pKa = 3.69GGGGADD711 pKa = 3.82TIFGGTGNDD720 pKa = 3.45TVSGGNGRR728 pKa = 11.84DD729 pKa = 3.71TVDD732 pKa = 3.79LGAGNDD738 pKa = 3.68VYY740 pKa = 11.33TDD742 pKa = 3.4TAQGDD747 pKa = 3.67FHH749 pKa = 9.04GEE751 pKa = 3.85DD752 pKa = 4.21TISGGAGNDD761 pKa = 3.82RR762 pKa = 11.84INGGGGNDD770 pKa = 4.66TINGDD775 pKa = 3.77NGSDD779 pKa = 3.55TLFGGAGDD787 pKa = 3.99DD788 pKa = 4.45TIHH791 pKa = 6.72GGAGVDD797 pKa = 3.65VLNGGTGADD806 pKa = 3.32ILTGGEE812 pKa = 4.09GADD815 pKa = 3.33TFVYY819 pKa = 10.26TSVGQSNFAGGIDD832 pKa = 4.06TITDD836 pKa = 4.07FTRR839 pKa = 11.84GTDD842 pKa = 3.42IIDD845 pKa = 4.14LSGIDD850 pKa = 4.15ANTSVAGNQAFGFIGGLDD868 pKa = 3.61FTGTAGEE875 pKa = 4.1LRR877 pKa = 11.84FANGQLLGDD886 pKa = 3.71VDD888 pKa = 4.48GDD890 pKa = 4.14GNSDD894 pKa = 3.48FSINLTGVGNLSGANLEE911 pKa = 4.49LGVDD915 pKa = 4.01EE916 pKa = 5.39EE917 pKa = 4.61PTDD920 pKa = 4.08GEE922 pKa = 4.57GGGGGGDD929 pKa = 3.74PVILPNDD936 pKa = 3.7NGQPAPEE943 pKa = 3.73QDD945 pKa = 3.36TFGEE949 pKa = 4.71SNPSLGMGLEE959 pKa = 5.39GIADD963 pKa = 3.93WSSQAPFIDD972 pKa = 3.34VMKK975 pKa = 10.34FARR978 pKa = 11.84PWIGHH983 pKa = 6.44VGDD986 pKa = 3.13QWGGYY991 pKa = 8.05GHH993 pKa = 7.58DD994 pKa = 3.55RR995 pKa = 11.84LEE997 pKa = 4.42EE998 pKa = 4.04NGFLDD1003 pKa = 4.12EE1004 pKa = 5.24NGWPTDD1010 pKa = 3.07IPAGVSSIQAVVMADD1025 pKa = 3.64LPEE1028 pKa = 4.49EE1029 pKa = 4.28SASLAGRR1036 pKa = 11.84YY1037 pKa = 9.17RR1038 pKa = 11.84LTWEE1042 pKa = 4.22GEE1044 pKa = 3.84GDD1046 pKa = 3.72VEE1048 pKa = 4.92VIGGRR1053 pKa = 11.84ISNITRR1059 pKa = 11.84SNGEE1063 pKa = 3.2IWFDD1067 pKa = 3.81FEE1069 pKa = 4.95PGGSGTVISISSTDD1083 pKa = 3.44PNNNGNYY1090 pKa = 9.22IRR1092 pKa = 11.84DD1093 pKa = 3.55IEE1095 pKa = 4.59LVHH1098 pKa = 6.79EE1099 pKa = 4.85DD1100 pKa = 3.54NIEE1103 pKa = 3.82AHH1105 pKa = 5.97EE1106 pKa = 4.73AGAIFNPDD1114 pKa = 3.77WIDD1117 pKa = 4.08LIEE1120 pKa = 4.7DD1121 pKa = 3.55LRR1123 pKa = 11.84SVRR1126 pKa = 11.84FMDD1129 pKa = 4.14WMQTNNSPQVDD1140 pKa = 3.31WSDD1143 pKa = 3.28RR1144 pKa = 11.84PTPDD1148 pKa = 2.62NYY1150 pKa = 10.68TYY1152 pKa = 11.01FDD1154 pKa = 3.36GGVPVEE1160 pKa = 5.07IMVQLANQIGADD1172 pKa = 3.28PWFNMPHH1179 pKa = 5.54QANDD1183 pKa = 2.8EE1184 pKa = 4.2YY1185 pKa = 10.7MRR1187 pKa = 11.84EE1188 pKa = 3.71FAEE1191 pKa = 4.3YY1192 pKa = 10.83VRR1194 pKa = 11.84DD1195 pKa = 3.85NLDD1198 pKa = 3.52PEE1200 pKa = 3.87LRR1202 pKa = 11.84AYY1204 pKa = 10.49VEE1206 pKa = 4.23YY1207 pKa = 11.21SNEE1210 pKa = 3.17VWNYY1214 pKa = 10.37GFEE1217 pKa = 4.14QAHH1220 pKa = 5.9WAAEE1224 pKa = 3.92QATARR1229 pKa = 11.84WGNAANDD1236 pKa = 4.26GGWMQFYY1243 pKa = 10.09GARR1246 pKa = 11.84AAEE1249 pKa = 4.15MAIIWDD1255 pKa = 3.96DD1256 pKa = 3.68VFGNQADD1263 pKa = 3.61DD1264 pKa = 3.49RR1265 pKa = 11.84VINVIGTHH1273 pKa = 5.5TGWPGLEE1280 pKa = 3.25QYY1282 pKa = 9.29MFNSEE1287 pKa = 3.68LWVRR1291 pKa = 11.84EE1292 pKa = 4.08TGNDD1296 pKa = 3.14APYY1299 pKa = 10.02TYY1301 pKa = 10.61FDD1303 pKa = 3.84AYY1305 pKa = 10.8AVTGYY1310 pKa = 10.63FGIEE1314 pKa = 4.16LGNSRR1319 pKa = 11.84AGEE1322 pKa = 3.87VLGWIEE1328 pKa = 4.4DD1329 pKa = 3.7SRR1331 pKa = 11.84NAAIADD1337 pKa = 3.74ANAQGLTGSARR1348 pKa = 11.84TRR1350 pKa = 11.84YY1351 pKa = 9.43IEE1353 pKa = 3.79EE1354 pKa = 3.94HH1355 pKa = 6.56QYY1357 pKa = 11.39DD1358 pKa = 3.8QASAIAAEE1366 pKa = 4.25DD1367 pKa = 3.78LRR1369 pKa = 11.84TGSLAEE1375 pKa = 4.89LIGEE1379 pKa = 4.73TYY1381 pKa = 10.42AYY1383 pKa = 10.42QSAAATAAGLEE1394 pKa = 4.07MVMYY1398 pKa = 10.48EE1399 pKa = 4.16GGTHH1403 pKa = 4.37VVGVGGNVNNAEE1415 pKa = 3.74LAAFFNHH1422 pKa = 6.3FNYY1425 pKa = 9.04TPEE1428 pKa = 3.82MAEE1431 pKa = 4.02LYY1433 pKa = 10.04EE1434 pKa = 4.35ILLEE1438 pKa = 4.3GWQAAGGGLFNAFVDD1453 pKa = 4.17VSDD1456 pKa = 4.3PSQYY1460 pKa = 11.11GSWGALRR1467 pKa = 11.84YY1468 pKa = 10.27LEE1470 pKa = 5.19DD1471 pKa = 5.65DD1472 pKa = 3.59NPRR1475 pKa = 11.84WDD1477 pKa = 3.88VLNDD1481 pKa = 3.29FNNNVAAWWEE1491 pKa = 4.09DD1492 pKa = 3.23RR1493 pKa = 11.84SADD1496 pKa = 3.7TFDD1499 pKa = 3.04QGVFINGTDD1508 pKa = 3.56GNDD1511 pKa = 3.99AIGGTAEE1518 pKa = 4.17EE1519 pKa = 5.51DD1520 pKa = 3.16ILLGGRR1526 pKa = 11.84GNDD1529 pKa = 3.29VLSGYY1534 pKa = 10.39GGEE1537 pKa = 4.42DD1538 pKa = 3.29RR1539 pKa = 11.84LHH1541 pKa = 7.72GGMGNDD1547 pKa = 3.87TAWLIGSYY1555 pKa = 10.29EE1556 pKa = 4.55DD1557 pKa = 3.75YY1558 pKa = 10.94TFTSDD1563 pKa = 3.05NGTVIATHH1571 pKa = 5.85SSGRR1575 pKa = 11.84TFLTDD1580 pKa = 3.04IEE1582 pKa = 4.31NVVFEE1587 pKa = 4.81EE1588 pKa = 4.4NPDD1591 pKa = 3.09VAYY1594 pKa = 10.26AANSLFF1600 pKa = 3.77
MM1 pKa = 7.27TEE3 pKa = 3.11IRR5 pKa = 11.84NFIFGNSLINHH16 pKa = 5.86VSGSHH21 pKa = 4.44QTTVPYY27 pKa = 10.08WLDD30 pKa = 3.32ALADD34 pKa = 4.16TAGHH38 pKa = 6.3SYY40 pKa = 11.01SVDD43 pKa = 3.24GQFGNLLSHH52 pKa = 7.28LGTNPIPNWWFGDD65 pKa = 3.78GVEE68 pKa = 4.68GSWNSSFANSDD79 pKa = 3.45YY80 pKa = 9.76TNVMITTANYY90 pKa = 9.5IQYY93 pKa = 9.93QGPDD97 pKa = 2.91EE98 pKa = 4.07QYY100 pKa = 10.28YY101 pKa = 10.51GNSHH105 pKa = 5.69TPISATLEE113 pKa = 4.1LVDD116 pKa = 3.86MVHH119 pKa = 6.82ARR121 pKa = 11.84EE122 pKa = 4.08PNAVVDD128 pKa = 5.43IYY130 pKa = 11.61VGWPDD135 pKa = 3.38MAGVFPNFPPSDD147 pKa = 3.85AQFDD151 pKa = 4.3TYY153 pKa = 11.61NQLTMTTYY161 pKa = 9.88MEE163 pKa = 4.61WYY165 pKa = 9.7DD166 pKa = 3.69EE167 pKa = 4.39YY168 pKa = 11.73YY169 pKa = 11.02AALQAARR176 pKa = 11.84PGVTMNLMPVGPAMAHH192 pKa = 6.89LFTTVLSDD200 pKa = 3.16VSAADD205 pKa = 4.05LYY207 pKa = 11.29EE208 pKa = 5.15DD209 pKa = 4.01NAPHH213 pKa = 5.76GTEE216 pKa = 3.66TLYY219 pKa = 11.13FLASLATYY227 pKa = 10.82AFMYY231 pKa = 10.57DD232 pKa = 4.04EE233 pKa = 5.55LPPADD238 pKa = 3.95FQIPNNILPSVRR250 pKa = 11.84AAYY253 pKa = 10.6SDD255 pKa = 3.4ILTEE259 pKa = 4.21LADD262 pKa = 3.79FTGIDD267 pKa = 3.53PAEE270 pKa = 4.17VEE272 pKa = 4.49GGSDD276 pKa = 3.81GDD278 pKa = 3.93SSGGGVDD285 pKa = 4.17VEE287 pKa = 4.57PEE289 pKa = 4.23VNVVDD294 pKa = 5.12GDD296 pKa = 3.86DD297 pKa = 3.56RR298 pKa = 11.84NNNIVGTDD306 pKa = 3.28GMDD309 pKa = 4.3AISGLGGNDD318 pKa = 3.56TIQAGDD324 pKa = 3.7GNDD327 pKa = 4.02TIDD330 pKa = 4.01AGAGNDD336 pKa = 4.78SVWGDD341 pKa = 3.39QGADD345 pKa = 3.62NIHH348 pKa = 6.54GGDD351 pKa = 3.58GSDD354 pKa = 3.55FLSGGRR360 pKa = 11.84GNDD363 pKa = 3.18VVEE366 pKa = 4.77GNSGDD371 pKa = 3.42DD372 pKa = 4.01WINGNYY378 pKa = 10.28GNDD381 pKa = 3.15ILNGGGGNDD390 pKa = 4.35TINAGAGRR398 pKa = 11.84DD399 pKa = 3.63TIYY402 pKa = 10.97AGNGDD407 pKa = 3.57DD408 pKa = 5.05RR409 pKa = 11.84VRR411 pKa = 11.84GGNGRR416 pKa = 11.84DD417 pKa = 3.38TVYY420 pKa = 10.88LGAGHH425 pKa = 7.51DD426 pKa = 3.48RR427 pKa = 11.84FTDD430 pKa = 3.33NGQTGEE436 pKa = 4.19NAVDD440 pKa = 3.49TVYY443 pKa = 11.21GGDD446 pKa = 3.77GDD448 pKa = 4.67DD449 pKa = 5.12AIHH452 pKa = 7.0GGGGDD457 pKa = 3.4DD458 pKa = 3.31RR459 pKa = 11.84FYY461 pKa = 11.74GEE463 pKa = 4.84GGNDD467 pKa = 3.67RR468 pKa = 11.84IFGGSGNDD476 pKa = 3.59QIRR479 pKa = 11.84GGAGDD484 pKa = 4.25DD485 pKa = 3.71SLHH488 pKa = 5.84GRR490 pKa = 11.84EE491 pKa = 4.23GADD494 pKa = 3.62TIFGGNGHH502 pKa = 6.8DD503 pKa = 4.82FINAGSGNDD512 pKa = 3.46TVEE515 pKa = 4.4GGNGRR520 pKa = 11.84DD521 pKa = 3.22RR522 pKa = 11.84VRR524 pKa = 11.84LGGGDD529 pKa = 4.09DD530 pKa = 3.48RR531 pKa = 11.84FVDD534 pKa = 3.43NDD536 pKa = 3.19QAGFQGSDD544 pKa = 3.29TVSGGAGNDD553 pKa = 3.77TILGGGGNDD562 pKa = 3.13RR563 pKa = 11.84LFGGSGADD571 pKa = 3.66TINGGLEE578 pKa = 3.78NDD580 pKa = 4.67HH581 pKa = 6.88ISGGEE586 pKa = 4.03GADD589 pKa = 3.55TLNGGWGDD597 pKa = 3.83DD598 pKa = 4.56FINGAQGADD607 pKa = 3.38IINAGGGDD615 pKa = 3.43DD616 pKa = 4.38HH617 pKa = 8.97VFGGLGADD625 pKa = 3.71VVDD628 pKa = 5.45LGDD631 pKa = 4.52GNDD634 pKa = 3.52IFQDD638 pKa = 3.59SAEE641 pKa = 4.18QGASGADD648 pKa = 3.36TVYY651 pKa = 11.05GGAGNDD657 pKa = 3.72IINGGGGQDD666 pKa = 3.67SLHH669 pKa = 6.67GGDD672 pKa = 4.65GDD674 pKa = 4.17DD675 pKa = 3.64EE676 pKa = 4.58VRR678 pKa = 11.84GGVGNDD684 pKa = 4.56DD685 pKa = 3.46ITGGAGNDD693 pKa = 3.73VLHH696 pKa = 6.92GGAGDD701 pKa = 3.96DD702 pKa = 4.77SIDD705 pKa = 3.69GGGGADD711 pKa = 3.82TIFGGTGNDD720 pKa = 3.45TVSGGNGRR728 pKa = 11.84DD729 pKa = 3.71TVDD732 pKa = 3.79LGAGNDD738 pKa = 3.68VYY740 pKa = 11.33TDD742 pKa = 3.4TAQGDD747 pKa = 3.67FHH749 pKa = 9.04GEE751 pKa = 3.85DD752 pKa = 4.21TISGGAGNDD761 pKa = 3.82RR762 pKa = 11.84INGGGGNDD770 pKa = 4.66TINGDD775 pKa = 3.77NGSDD779 pKa = 3.55TLFGGAGDD787 pKa = 3.99DD788 pKa = 4.45TIHH791 pKa = 6.72GGAGVDD797 pKa = 3.65VLNGGTGADD806 pKa = 3.32ILTGGEE812 pKa = 4.09GADD815 pKa = 3.33TFVYY819 pKa = 10.26TSVGQSNFAGGIDD832 pKa = 4.06TITDD836 pKa = 4.07FTRR839 pKa = 11.84GTDD842 pKa = 3.42IIDD845 pKa = 4.14LSGIDD850 pKa = 4.15ANTSVAGNQAFGFIGGLDD868 pKa = 3.61FTGTAGEE875 pKa = 4.1LRR877 pKa = 11.84FANGQLLGDD886 pKa = 3.71VDD888 pKa = 4.48GDD890 pKa = 4.14GNSDD894 pKa = 3.48FSINLTGVGNLSGANLEE911 pKa = 4.49LGVDD915 pKa = 4.01EE916 pKa = 5.39EE917 pKa = 4.61PTDD920 pKa = 4.08GEE922 pKa = 4.57GGGGGGDD929 pKa = 3.74PVILPNDD936 pKa = 3.7NGQPAPEE943 pKa = 3.73QDD945 pKa = 3.36TFGEE949 pKa = 4.71SNPSLGMGLEE959 pKa = 5.39GIADD963 pKa = 3.93WSSQAPFIDD972 pKa = 3.34VMKK975 pKa = 10.34FARR978 pKa = 11.84PWIGHH983 pKa = 6.44VGDD986 pKa = 3.13QWGGYY991 pKa = 8.05GHH993 pKa = 7.58DD994 pKa = 3.55RR995 pKa = 11.84LEE997 pKa = 4.42EE998 pKa = 4.04NGFLDD1003 pKa = 4.12EE1004 pKa = 5.24NGWPTDD1010 pKa = 3.07IPAGVSSIQAVVMADD1025 pKa = 3.64LPEE1028 pKa = 4.49EE1029 pKa = 4.28SASLAGRR1036 pKa = 11.84YY1037 pKa = 9.17RR1038 pKa = 11.84LTWEE1042 pKa = 4.22GEE1044 pKa = 3.84GDD1046 pKa = 3.72VEE1048 pKa = 4.92VIGGRR1053 pKa = 11.84ISNITRR1059 pKa = 11.84SNGEE1063 pKa = 3.2IWFDD1067 pKa = 3.81FEE1069 pKa = 4.95PGGSGTVISISSTDD1083 pKa = 3.44PNNNGNYY1090 pKa = 9.22IRR1092 pKa = 11.84DD1093 pKa = 3.55IEE1095 pKa = 4.59LVHH1098 pKa = 6.79EE1099 pKa = 4.85DD1100 pKa = 3.54NIEE1103 pKa = 3.82AHH1105 pKa = 5.97EE1106 pKa = 4.73AGAIFNPDD1114 pKa = 3.77WIDD1117 pKa = 4.08LIEE1120 pKa = 4.7DD1121 pKa = 3.55LRR1123 pKa = 11.84SVRR1126 pKa = 11.84FMDD1129 pKa = 4.14WMQTNNSPQVDD1140 pKa = 3.31WSDD1143 pKa = 3.28RR1144 pKa = 11.84PTPDD1148 pKa = 2.62NYY1150 pKa = 10.68TYY1152 pKa = 11.01FDD1154 pKa = 3.36GGVPVEE1160 pKa = 5.07IMVQLANQIGADD1172 pKa = 3.28PWFNMPHH1179 pKa = 5.54QANDD1183 pKa = 2.8EE1184 pKa = 4.2YY1185 pKa = 10.7MRR1187 pKa = 11.84EE1188 pKa = 3.71FAEE1191 pKa = 4.3YY1192 pKa = 10.83VRR1194 pKa = 11.84DD1195 pKa = 3.85NLDD1198 pKa = 3.52PEE1200 pKa = 3.87LRR1202 pKa = 11.84AYY1204 pKa = 10.49VEE1206 pKa = 4.23YY1207 pKa = 11.21SNEE1210 pKa = 3.17VWNYY1214 pKa = 10.37GFEE1217 pKa = 4.14QAHH1220 pKa = 5.9WAAEE1224 pKa = 3.92QATARR1229 pKa = 11.84WGNAANDD1236 pKa = 4.26GGWMQFYY1243 pKa = 10.09GARR1246 pKa = 11.84AAEE1249 pKa = 4.15MAIIWDD1255 pKa = 3.96DD1256 pKa = 3.68VFGNQADD1263 pKa = 3.61DD1264 pKa = 3.49RR1265 pKa = 11.84VINVIGTHH1273 pKa = 5.5TGWPGLEE1280 pKa = 3.25QYY1282 pKa = 9.29MFNSEE1287 pKa = 3.68LWVRR1291 pKa = 11.84EE1292 pKa = 4.08TGNDD1296 pKa = 3.14APYY1299 pKa = 10.02TYY1301 pKa = 10.61FDD1303 pKa = 3.84AYY1305 pKa = 10.8AVTGYY1310 pKa = 10.63FGIEE1314 pKa = 4.16LGNSRR1319 pKa = 11.84AGEE1322 pKa = 3.87VLGWIEE1328 pKa = 4.4DD1329 pKa = 3.7SRR1331 pKa = 11.84NAAIADD1337 pKa = 3.74ANAQGLTGSARR1348 pKa = 11.84TRR1350 pKa = 11.84YY1351 pKa = 9.43IEE1353 pKa = 3.79EE1354 pKa = 3.94HH1355 pKa = 6.56QYY1357 pKa = 11.39DD1358 pKa = 3.8QASAIAAEE1366 pKa = 4.25DD1367 pKa = 3.78LRR1369 pKa = 11.84TGSLAEE1375 pKa = 4.89LIGEE1379 pKa = 4.73TYY1381 pKa = 10.42AYY1383 pKa = 10.42QSAAATAAGLEE1394 pKa = 4.07MVMYY1398 pKa = 10.48EE1399 pKa = 4.16GGTHH1403 pKa = 4.37VVGVGGNVNNAEE1415 pKa = 3.74LAAFFNHH1422 pKa = 6.3FNYY1425 pKa = 9.04TPEE1428 pKa = 3.82MAEE1431 pKa = 4.02LYY1433 pKa = 10.04EE1434 pKa = 4.35ILLEE1438 pKa = 4.3GWQAAGGGLFNAFVDD1453 pKa = 4.17VSDD1456 pKa = 4.3PSQYY1460 pKa = 11.11GSWGALRR1467 pKa = 11.84YY1468 pKa = 10.27LEE1470 pKa = 5.19DD1471 pKa = 5.65DD1472 pKa = 3.59NPRR1475 pKa = 11.84WDD1477 pKa = 3.88VLNDD1481 pKa = 3.29FNNNVAAWWEE1491 pKa = 4.09DD1492 pKa = 3.23RR1493 pKa = 11.84SADD1496 pKa = 3.7TFDD1499 pKa = 3.04QGVFINGTDD1508 pKa = 3.56GNDD1511 pKa = 3.99AIGGTAEE1518 pKa = 4.17EE1519 pKa = 5.51DD1520 pKa = 3.16ILLGGRR1526 pKa = 11.84GNDD1529 pKa = 3.29VLSGYY1534 pKa = 10.39GGEE1537 pKa = 4.42DD1538 pKa = 3.29RR1539 pKa = 11.84LHH1541 pKa = 7.72GGMGNDD1547 pKa = 3.87TAWLIGSYY1555 pKa = 10.29EE1556 pKa = 4.55DD1557 pKa = 3.75YY1558 pKa = 10.94TFTSDD1563 pKa = 3.05NGTVIATHH1571 pKa = 5.85SSGRR1575 pKa = 11.84TFLTDD1580 pKa = 3.04IEE1582 pKa = 4.31NVVFEE1587 pKa = 4.81EE1588 pKa = 4.4NPDD1591 pKa = 3.09VAYY1594 pKa = 10.26AANSLFF1600 pKa = 3.77
Molecular weight: 167.87 kDa
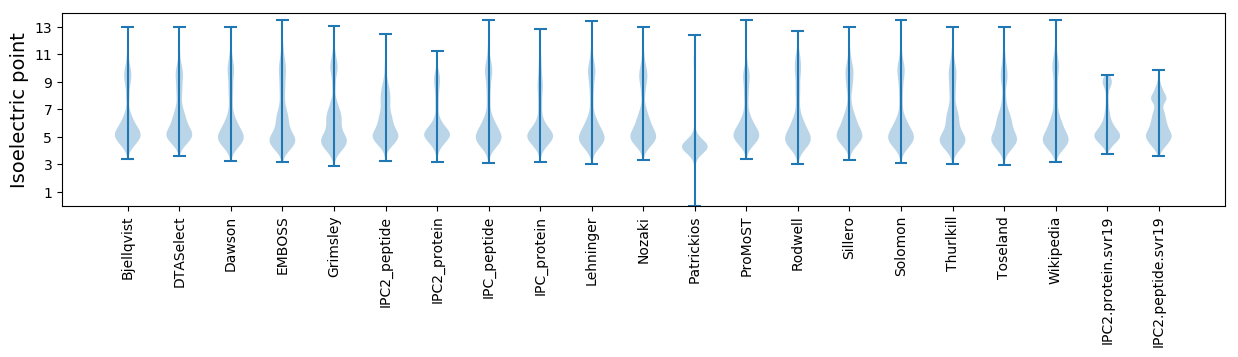
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P1SZC8|A0A6P1SZC8_9RHOB Cysteine desulfurase OS=Rhodobacteraceae bacterium 9Alg 56 OX=2683284 GN=GO499_06215 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.78ILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3SLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.78ILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3SLSAA44 pKa = 3.83
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1205218 |
31 |
2168 |
313.6 |
34.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.302 ± 0.05 | 0.899 ± 0.013 |
5.819 ± 0.033 | 6.339 ± 0.035 |
3.873 ± 0.025 | 8.775 ± 0.044 |
2.006 ± 0.02 | 5.209 ± 0.031 |
2.951 ± 0.034 | 10.061 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.68 ± 0.02 | 2.623 ± 0.021 |
5.069 ± 0.03 | 2.973 ± 0.02 |
6.779 ± 0.037 | 5.34 ± 0.025 |
5.498 ± 0.028 | 7.242 ± 0.032 |
1.365 ± 0.017 | 2.196 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
