
Planococcus sp. PAMC 21323
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Planococcaceae; Planococcus; unclassified Planococcus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
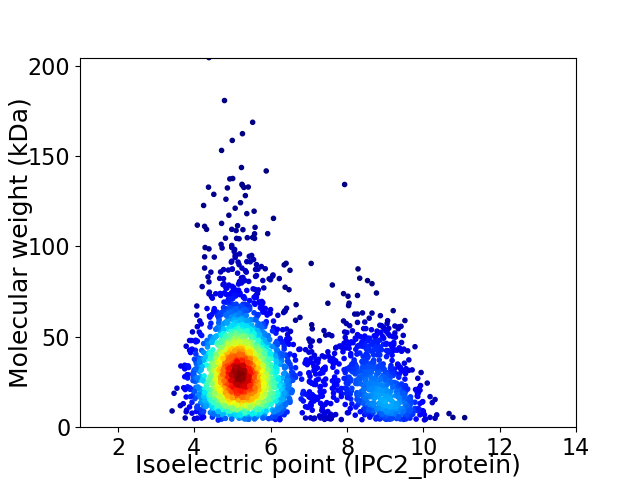
Virtual 2D-PAGE plot for 3084 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4RDN9|A0A0B4RDN9_9BACL Uncharacterized protein OS=Planococcus sp. PAMC 21323 OX=1526927 GN=Plano_2314 PE=4 SV=1
MM1 pKa = 7.2TRR3 pKa = 11.84RR4 pKa = 11.84TSKK7 pKa = 10.42EE8 pKa = 3.53IKK10 pKa = 8.95KK11 pKa = 8.48TNQIIAALSIALVLVMALGIYY32 pKa = 10.82LMMKK36 pKa = 9.62PDD38 pKa = 4.39LPAEE42 pKa = 4.14TRR44 pKa = 11.84PPQSTQQQTEE54 pKa = 4.1SGTPEE59 pKa = 4.38AEE61 pKa = 4.05TEE63 pKa = 4.15LASMIANAKK72 pKa = 5.94THH74 pKa = 6.26VYY76 pKa = 9.01TLYY79 pKa = 10.62TDD81 pKa = 4.87LEE83 pKa = 4.35QGSGFLINNQGDD95 pKa = 3.95ILTNAHH101 pKa = 5.65VVLDD105 pKa = 3.96ASYY108 pKa = 8.65ITVKK112 pKa = 10.86NSDD115 pKa = 3.36GQEE118 pKa = 4.04FNGNLIGISEE128 pKa = 4.39TQDD131 pKa = 3.37LAIVRR136 pKa = 11.84VSEE139 pKa = 4.18LAGKK143 pKa = 9.74EE144 pKa = 3.74PLEE147 pKa = 4.66IEE149 pKa = 4.44MEE151 pKa = 4.21PVDD154 pKa = 3.24IGTPVVAIGSPNDD167 pKa = 3.43QSNTATIGEE176 pKa = 4.28ITDD179 pKa = 3.79TEE181 pKa = 5.05LDD183 pKa = 3.56FSDD186 pKa = 3.23QFEE189 pKa = 4.51YY190 pKa = 10.94TNLYY194 pKa = 9.79EE195 pKa = 4.16MSAEE199 pKa = 3.9IAQGSSGGPLIAADD213 pKa = 3.68TGKK216 pKa = 10.77LLGINSIIIEE226 pKa = 4.27EE227 pKa = 4.24NPDD230 pKa = 3.2LGYY233 pKa = 10.65AIPIYY238 pKa = 8.4TVWDD242 pKa = 3.63QLTEE246 pKa = 4.58WINDD250 pKa = 3.77PLITEE255 pKa = 4.13EE256 pKa = 4.48EE257 pKa = 4.51EE258 pKa = 4.96IVLQDD263 pKa = 3.55VKK265 pKa = 11.03DD266 pKa = 3.81AYY268 pKa = 10.92FDD270 pKa = 3.82EE271 pKa = 5.87DD272 pKa = 3.89FLAGFISAYY281 pKa = 10.65YY282 pKa = 9.92EE283 pKa = 3.93LLPYY287 pKa = 10.39SLNDD291 pKa = 3.65RR292 pKa = 11.84EE293 pKa = 4.46STYY296 pKa = 10.97YY297 pKa = 9.52MSYY300 pKa = 10.53IFPGSNAEE308 pKa = 4.0QQATGLIEE316 pKa = 4.7EE317 pKa = 4.64YY318 pKa = 10.7SDD320 pKa = 3.73PNLIYY325 pKa = 10.75DD326 pKa = 3.8VVKK329 pKa = 8.98PTITSIEE336 pKa = 4.01IEE338 pKa = 4.17GDD340 pKa = 3.55SALVEE345 pKa = 4.26AEE347 pKa = 4.68AEE349 pKa = 4.1FTYY352 pKa = 10.6HH353 pKa = 7.54DD354 pKa = 4.05KK355 pKa = 11.48ASDD358 pKa = 3.34KK359 pKa = 11.23SSTISHH365 pKa = 6.62KK366 pKa = 10.87GMYY369 pKa = 8.65TVVIDD374 pKa = 4.44EE375 pKa = 4.51YY376 pKa = 11.55GDD378 pKa = 3.67YY379 pKa = 10.99QIDD382 pKa = 4.56GIVNQQ387 pKa = 4.23
MM1 pKa = 7.2TRR3 pKa = 11.84RR4 pKa = 11.84TSKK7 pKa = 10.42EE8 pKa = 3.53IKK10 pKa = 8.95KK11 pKa = 8.48TNQIIAALSIALVLVMALGIYY32 pKa = 10.82LMMKK36 pKa = 9.62PDD38 pKa = 4.39LPAEE42 pKa = 4.14TRR44 pKa = 11.84PPQSTQQQTEE54 pKa = 4.1SGTPEE59 pKa = 4.38AEE61 pKa = 4.05TEE63 pKa = 4.15LASMIANAKK72 pKa = 5.94THH74 pKa = 6.26VYY76 pKa = 9.01TLYY79 pKa = 10.62TDD81 pKa = 4.87LEE83 pKa = 4.35QGSGFLINNQGDD95 pKa = 3.95ILTNAHH101 pKa = 5.65VVLDD105 pKa = 3.96ASYY108 pKa = 8.65ITVKK112 pKa = 10.86NSDD115 pKa = 3.36GQEE118 pKa = 4.04FNGNLIGISEE128 pKa = 4.39TQDD131 pKa = 3.37LAIVRR136 pKa = 11.84VSEE139 pKa = 4.18LAGKK143 pKa = 9.74EE144 pKa = 3.74PLEE147 pKa = 4.66IEE149 pKa = 4.44MEE151 pKa = 4.21PVDD154 pKa = 3.24IGTPVVAIGSPNDD167 pKa = 3.43QSNTATIGEE176 pKa = 4.28ITDD179 pKa = 3.79TEE181 pKa = 5.05LDD183 pKa = 3.56FSDD186 pKa = 3.23QFEE189 pKa = 4.51YY190 pKa = 10.94TNLYY194 pKa = 9.79EE195 pKa = 4.16MSAEE199 pKa = 3.9IAQGSSGGPLIAADD213 pKa = 3.68TGKK216 pKa = 10.77LLGINSIIIEE226 pKa = 4.27EE227 pKa = 4.24NPDD230 pKa = 3.2LGYY233 pKa = 10.65AIPIYY238 pKa = 8.4TVWDD242 pKa = 3.63QLTEE246 pKa = 4.58WINDD250 pKa = 3.77PLITEE255 pKa = 4.13EE256 pKa = 4.48EE257 pKa = 4.51EE258 pKa = 4.96IVLQDD263 pKa = 3.55VKK265 pKa = 11.03DD266 pKa = 3.81AYY268 pKa = 10.92FDD270 pKa = 3.82EE271 pKa = 5.87DD272 pKa = 3.89FLAGFISAYY281 pKa = 10.65YY282 pKa = 9.92EE283 pKa = 3.93LLPYY287 pKa = 10.39SLNDD291 pKa = 3.65RR292 pKa = 11.84EE293 pKa = 4.46STYY296 pKa = 10.97YY297 pKa = 9.52MSYY300 pKa = 10.53IFPGSNAEE308 pKa = 4.0QQATGLIEE316 pKa = 4.7EE317 pKa = 4.64YY318 pKa = 10.7SDD320 pKa = 3.73PNLIYY325 pKa = 10.75DD326 pKa = 3.8VVKK329 pKa = 8.98PTITSIEE336 pKa = 4.01IEE338 pKa = 4.17GDD340 pKa = 3.55SALVEE345 pKa = 4.26AEE347 pKa = 4.68AEE349 pKa = 4.1FTYY352 pKa = 10.6HH353 pKa = 7.54DD354 pKa = 4.05KK355 pKa = 11.48ASDD358 pKa = 3.34KK359 pKa = 11.23SSTISHH365 pKa = 6.62KK366 pKa = 10.87GMYY369 pKa = 8.65TVVIDD374 pKa = 4.44EE375 pKa = 4.51YY376 pKa = 11.55GDD378 pKa = 3.67YY379 pKa = 10.99QIDD382 pKa = 4.56GIVNQQ387 pKa = 4.23
Molecular weight: 42.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4RGE3|A0A0B4RGE3_9BACL Anthranilate synthase amidotransferase component OS=Planococcus sp. PAMC 21323 OX=1526927 GN=Plano_2877 PE=4 SV=1
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNTRR11 pKa = 11.84KK12 pKa = 9.66HH13 pKa = 5.87SKK15 pKa = 8.83VHH17 pKa = 5.7GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.52NGRR29 pKa = 11.84RR30 pKa = 11.84IIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.83GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNTRR11 pKa = 11.84KK12 pKa = 9.66HH13 pKa = 5.87SKK15 pKa = 8.83VHH17 pKa = 5.7GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.52NGRR29 pKa = 11.84RR30 pKa = 11.84IIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.83GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
915920 |
37 |
1846 |
297.0 |
33.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.696 ± 0.048 | 0.563 ± 0.013 |
5.243 ± 0.038 | 7.705 ± 0.055 |
4.598 ± 0.033 | 6.93 ± 0.047 |
1.997 ± 0.02 | 7.534 ± 0.04 |
6.336 ± 0.042 | 9.823 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.92 ± 0.024 | 4.051 ± 0.033 |
3.6 ± 0.019 | 3.822 ± 0.03 |
4.003 ± 0.028 | 6.067 ± 0.03 |
5.643 ± 0.028 | 7.167 ± 0.034 |
1.0 ± 0.016 | 3.3 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
