
Leopards Hill virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Nairoviridae; Orthonairovirus; Leopards Hill orthonairovirus
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
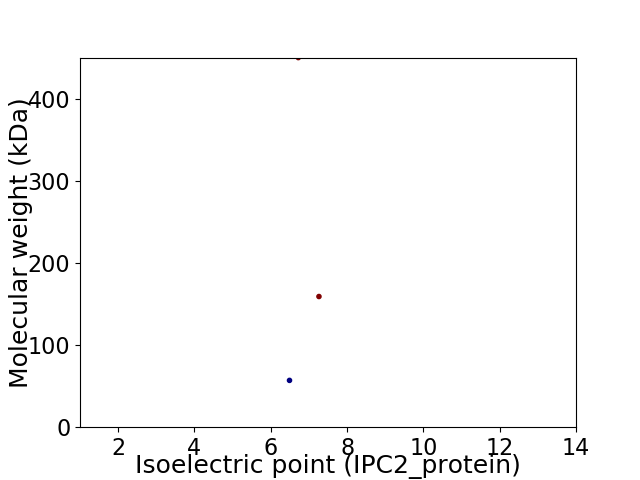
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B6VMT2|A0A0B6VMT2_9VIRU Glycoprotein G2 OS=Leopards Hill virus OX=1381104 GN=GP PE=4 SV=1
MM1 pKa = 7.75SEE3 pKa = 5.54LNFKK7 pKa = 10.19DD8 pKa = 5.12LNGLNNWFEE17 pKa = 4.22EE18 pKa = 4.45FKK20 pKa = 10.86KK21 pKa = 10.61DD22 pKa = 3.66VPIYY26 pKa = 10.16TSGANSACLSSGLVDD41 pKa = 4.33LSQFKK46 pKa = 11.23LEE48 pKa = 4.26IDD50 pKa = 3.68AQSSEE55 pKa = 4.28SAKK58 pKa = 9.75NAVYY62 pKa = 10.4GRR64 pKa = 11.84AVIAATRR71 pKa = 11.84NLAPIYY77 pKa = 8.7EE78 pKa = 4.69CAWTSCEE85 pKa = 4.32GIFEE89 pKa = 5.2RR90 pKa = 11.84SMSWFNDD97 pKa = 3.16NADD100 pKa = 3.59KK101 pKa = 11.3PEE103 pKa = 3.99FKK105 pKa = 10.26VWHH108 pKa = 6.95DD109 pKa = 3.85KK110 pKa = 11.64YY111 pKa = 11.71NDD113 pKa = 3.62LKK115 pKa = 11.51SNLPEE120 pKa = 3.93VEE122 pKa = 4.63QILAYY127 pKa = 9.3QNCSSLWRR135 pKa = 11.84KK136 pKa = 10.24DD137 pKa = 2.69IGYY140 pKa = 9.09EE141 pKa = 3.84INQNTSTLKK150 pKa = 10.08GDD152 pKa = 3.19IVEE155 pKa = 4.39EE156 pKa = 4.21YY157 pKa = 10.57GVNRR161 pKa = 11.84NIVGTVKK168 pKa = 10.74NMLVDD173 pKa = 3.66MVNKK177 pKa = 10.01RR178 pKa = 11.84KK179 pKa = 9.86KK180 pKa = 9.56RR181 pKa = 11.84LGGVVITGAGTSEE194 pKa = 4.55DD195 pKa = 4.04HH196 pKa = 6.02ITWMEE201 pKa = 3.49NWLDD205 pKa = 3.69GEE207 pKa = 4.56IEE209 pKa = 4.34SFSIPGWGSWAKK221 pKa = 10.38QNSKK225 pKa = 8.16GTRR228 pKa = 11.84LAGTAIANIMQKK240 pKa = 10.23KK241 pKa = 10.53DD242 pKa = 2.96SDD244 pKa = 3.94ALEE247 pKa = 4.15KK248 pKa = 10.91AKK250 pKa = 10.91EE251 pKa = 4.13KK252 pKa = 10.66LEE254 pKa = 4.05KK255 pKa = 9.58MKK257 pKa = 9.63KK258 pKa = 7.73TADD261 pKa = 3.67DD262 pKa = 4.01PASLEE267 pKa = 4.2SEE269 pKa = 4.28GMSEE273 pKa = 4.22DD274 pKa = 3.68AAKK277 pKa = 10.59KK278 pKa = 8.56MCQEE282 pKa = 3.96VEE284 pKa = 3.9ASIEE288 pKa = 4.01EE289 pKa = 4.33AEE291 pKa = 4.41VLLGTNVAQGEE302 pKa = 4.61SKK304 pKa = 10.71YY305 pKa = 9.53QQQAAAMDD313 pKa = 4.24VPFSAHH319 pKa = 3.7YY320 pKa = 8.1WAWRR324 pKa = 11.84CKK326 pKa = 9.9VNEE329 pKa = 4.01NSFPALSQWLFEE341 pKa = 4.58LGQRR345 pKa = 11.84PIGAAKK351 pKa = 9.72INTMLGEE358 pKa = 4.77LPFIWARR365 pKa = 11.84NIRR368 pKa = 11.84GSFASSNFNGNKK380 pKa = 9.33IYY382 pKa = 8.85MHH384 pKa = 6.78PAVLTAGRR392 pKa = 11.84LSDD395 pKa = 3.39MAACFGAFPVANPQRR410 pKa = 11.84AAEE413 pKa = 4.3GTGNTRR419 pKa = 11.84YY420 pKa = 10.31LLNLKK425 pKa = 10.41RR426 pKa = 11.84EE427 pKa = 4.03GDD429 pKa = 3.88NPCASMVTSLFDD441 pKa = 3.3VFSAGFKK448 pKa = 9.06YY449 pKa = 10.58QDD451 pKa = 3.41MDD453 pKa = 3.67IVPPEE458 pKa = 4.16HH459 pKa = 6.65MLHH462 pKa = 6.36QSFLGKK468 pKa = 9.1TSPFQTANKK477 pKa = 9.66IKK479 pKa = 10.79GSFTKK484 pKa = 10.25INVVAAPNKK493 pKa = 5.83PTPYY497 pKa = 10.39VFTPASAKK505 pKa = 10.08KK506 pKa = 9.97IKK508 pKa = 9.73EE509 pKa = 3.89KK510 pKa = 10.43KK511 pKa = 9.15ARR513 pKa = 11.84KK514 pKa = 8.96
MM1 pKa = 7.75SEE3 pKa = 5.54LNFKK7 pKa = 10.19DD8 pKa = 5.12LNGLNNWFEE17 pKa = 4.22EE18 pKa = 4.45FKK20 pKa = 10.86KK21 pKa = 10.61DD22 pKa = 3.66VPIYY26 pKa = 10.16TSGANSACLSSGLVDD41 pKa = 4.33LSQFKK46 pKa = 11.23LEE48 pKa = 4.26IDD50 pKa = 3.68AQSSEE55 pKa = 4.28SAKK58 pKa = 9.75NAVYY62 pKa = 10.4GRR64 pKa = 11.84AVIAATRR71 pKa = 11.84NLAPIYY77 pKa = 8.7EE78 pKa = 4.69CAWTSCEE85 pKa = 4.32GIFEE89 pKa = 5.2RR90 pKa = 11.84SMSWFNDD97 pKa = 3.16NADD100 pKa = 3.59KK101 pKa = 11.3PEE103 pKa = 3.99FKK105 pKa = 10.26VWHH108 pKa = 6.95DD109 pKa = 3.85KK110 pKa = 11.64YY111 pKa = 11.71NDD113 pKa = 3.62LKK115 pKa = 11.51SNLPEE120 pKa = 3.93VEE122 pKa = 4.63QILAYY127 pKa = 9.3QNCSSLWRR135 pKa = 11.84KK136 pKa = 10.24DD137 pKa = 2.69IGYY140 pKa = 9.09EE141 pKa = 3.84INQNTSTLKK150 pKa = 10.08GDD152 pKa = 3.19IVEE155 pKa = 4.39EE156 pKa = 4.21YY157 pKa = 10.57GVNRR161 pKa = 11.84NIVGTVKK168 pKa = 10.74NMLVDD173 pKa = 3.66MVNKK177 pKa = 10.01RR178 pKa = 11.84KK179 pKa = 9.86KK180 pKa = 9.56RR181 pKa = 11.84LGGVVITGAGTSEE194 pKa = 4.55DD195 pKa = 4.04HH196 pKa = 6.02ITWMEE201 pKa = 3.49NWLDD205 pKa = 3.69GEE207 pKa = 4.56IEE209 pKa = 4.34SFSIPGWGSWAKK221 pKa = 10.38QNSKK225 pKa = 8.16GTRR228 pKa = 11.84LAGTAIANIMQKK240 pKa = 10.23KK241 pKa = 10.53DD242 pKa = 2.96SDD244 pKa = 3.94ALEE247 pKa = 4.15KK248 pKa = 10.91AKK250 pKa = 10.91EE251 pKa = 4.13KK252 pKa = 10.66LEE254 pKa = 4.05KK255 pKa = 9.58MKK257 pKa = 9.63KK258 pKa = 7.73TADD261 pKa = 3.67DD262 pKa = 4.01PASLEE267 pKa = 4.2SEE269 pKa = 4.28GMSEE273 pKa = 4.22DD274 pKa = 3.68AAKK277 pKa = 10.59KK278 pKa = 8.56MCQEE282 pKa = 3.96VEE284 pKa = 3.9ASIEE288 pKa = 4.01EE289 pKa = 4.33AEE291 pKa = 4.41VLLGTNVAQGEE302 pKa = 4.61SKK304 pKa = 10.71YY305 pKa = 9.53QQQAAAMDD313 pKa = 4.24VPFSAHH319 pKa = 3.7YY320 pKa = 8.1WAWRR324 pKa = 11.84CKK326 pKa = 9.9VNEE329 pKa = 4.01NSFPALSQWLFEE341 pKa = 4.58LGQRR345 pKa = 11.84PIGAAKK351 pKa = 9.72INTMLGEE358 pKa = 4.77LPFIWARR365 pKa = 11.84NIRR368 pKa = 11.84GSFASSNFNGNKK380 pKa = 9.33IYY382 pKa = 8.85MHH384 pKa = 6.78PAVLTAGRR392 pKa = 11.84LSDD395 pKa = 3.39MAACFGAFPVANPQRR410 pKa = 11.84AAEE413 pKa = 4.3GTGNTRR419 pKa = 11.84YY420 pKa = 10.31LLNLKK425 pKa = 10.41RR426 pKa = 11.84EE427 pKa = 4.03GDD429 pKa = 3.88NPCASMVTSLFDD441 pKa = 3.3VFSAGFKK448 pKa = 9.06YY449 pKa = 10.58QDD451 pKa = 3.41MDD453 pKa = 3.67IVPPEE458 pKa = 4.16HH459 pKa = 6.65MLHH462 pKa = 6.36QSFLGKK468 pKa = 9.1TSPFQTANKK477 pKa = 9.66IKK479 pKa = 10.79GSFTKK484 pKa = 10.25INVVAAPNKK493 pKa = 5.83PTPYY497 pKa = 10.39VFTPASAKK505 pKa = 10.08KK506 pKa = 9.97IKK508 pKa = 9.73EE509 pKa = 3.89KK510 pKa = 10.43KK511 pKa = 9.15ARR513 pKa = 11.84KK514 pKa = 8.96
Molecular weight: 57.09 kDa
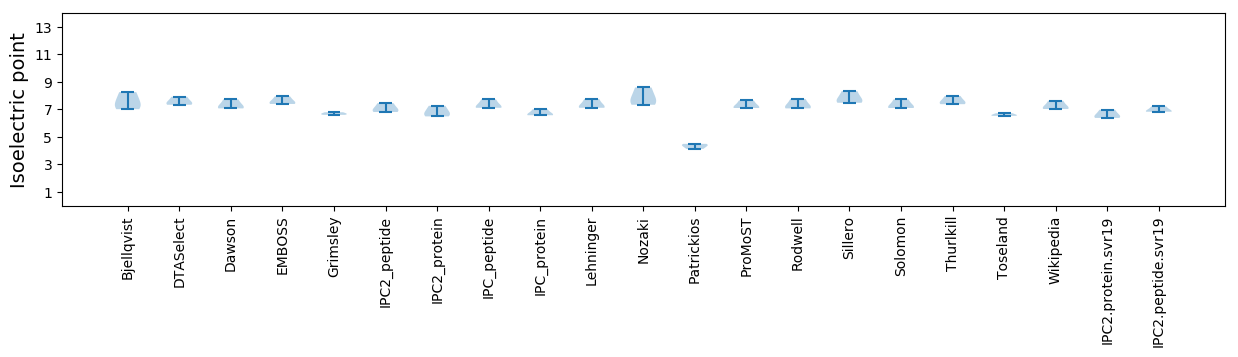
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B6VMT2|A0A0B6VMT2_9VIRU Glycoprotein G2 OS=Leopards Hill virus OX=1381104 GN=GP PE=4 SV=1
MM1 pKa = 6.81EE2 pKa = 5.24TYY4 pKa = 10.29HH5 pKa = 6.68GSRR8 pKa = 11.84YY9 pKa = 7.43TATSCLFFVIMLVVAITADD28 pKa = 3.58NNVTEE33 pKa = 4.43TTTAEE38 pKa = 4.36VVSNTTGSTEE48 pKa = 4.16GDD50 pKa = 3.67TTSKK54 pKa = 11.15GMDD57 pKa = 3.27TTQIPQINQTSATTRR72 pKa = 11.84SNNITTKK79 pKa = 9.21TSRR82 pKa = 11.84SEE84 pKa = 4.18SGVDD88 pKa = 3.64PSATQPLLVSSQASGTIGQQRR109 pKa = 11.84GLSPTPDD116 pKa = 3.28PGARR120 pKa = 11.84IRR122 pKa = 11.84VKK124 pKa = 10.11GGKK127 pKa = 8.98RR128 pKa = 11.84RR129 pKa = 11.84AEE131 pKa = 3.81RR132 pKa = 11.84LTARR136 pKa = 11.84LDD138 pKa = 3.6VNGNRR143 pKa = 11.84VVLKK147 pKa = 10.78RR148 pKa = 11.84EE149 pKa = 3.59ISGNLQLAKK158 pKa = 10.22EE159 pKa = 4.05QLEE162 pKa = 4.18EE163 pKa = 3.97AVLNMEE169 pKa = 4.75IDD171 pKa = 3.86SLDD174 pKa = 3.73LEE176 pKa = 4.35QLEE179 pKa = 4.57EE180 pKa = 3.97EE181 pKa = 4.29SPYY184 pKa = 11.09RR185 pKa = 11.84SMQRR189 pKa = 11.84TRR191 pKa = 11.84LMLPKK196 pKa = 10.3LEE198 pKa = 4.03SFAYY202 pKa = 10.47NEE204 pKa = 4.07EE205 pKa = 4.03QEE207 pKa = 4.25EE208 pKa = 4.36DD209 pKa = 3.92VMVTGKK215 pKa = 10.62LSRR218 pKa = 11.84FTRR221 pKa = 11.84FEE223 pKa = 4.43RR224 pKa = 11.84INIDD228 pKa = 3.15KK229 pKa = 10.84LEE231 pKa = 4.79SITVFQNLTNRR242 pKa = 11.84SVLTLCTYY250 pKa = 10.47FGHH253 pKa = 6.63YY254 pKa = 9.23YY255 pKa = 10.93NKK257 pKa = 8.65TLKK260 pKa = 9.1GTTVSIHH267 pKa = 6.1HH268 pKa = 6.64KK269 pKa = 10.52CDD271 pKa = 3.71IEE273 pKa = 4.48TQCKK277 pKa = 10.05GLTEE281 pKa = 4.23HH282 pKa = 6.75VSIDD286 pKa = 4.48RR287 pKa = 11.84ITNYY291 pKa = 11.17DD292 pKa = 3.6NIKK295 pKa = 9.9IINIMEE301 pKa = 4.29LPVIIIAYY309 pKa = 5.52FTRR312 pKa = 11.84HH313 pKa = 6.03FGFTYY318 pKa = 10.11KK319 pKa = 10.71NLVIHH324 pKa = 6.14EE325 pKa = 4.31QIEE328 pKa = 4.41NCLFFYY334 pKa = 9.82PQNPVACMEE343 pKa = 4.41NNWGGRR349 pKa = 11.84SHH351 pKa = 7.13PVVHH355 pKa = 6.57LVIEE359 pKa = 4.62RR360 pKa = 11.84KK361 pKa = 9.12YY362 pKa = 11.01LKK364 pKa = 10.07ADD366 pKa = 3.55SSITLCGLKK375 pKa = 10.18KK376 pKa = 10.78GNPTSLKK383 pKa = 9.01GTWFAEE389 pKa = 4.3KK390 pKa = 10.06TKK392 pKa = 10.04VTIDD396 pKa = 3.12LGEE399 pKa = 4.21PQTAGRR405 pKa = 11.84RR406 pKa = 11.84KK407 pKa = 9.71LLAIEE412 pKa = 4.09RR413 pKa = 11.84RR414 pKa = 11.84RR415 pKa = 11.84RR416 pKa = 11.84LPGRR420 pKa = 11.84EE421 pKa = 4.51DD422 pKa = 3.21IRR424 pKa = 11.84CHH426 pKa = 6.73SGSHH430 pKa = 5.66LVKK433 pKa = 9.49IDD435 pKa = 3.31KK436 pKa = 9.64YY437 pKa = 11.14APTNEE442 pKa = 4.08FQSFPNAKK450 pKa = 9.55IGFCNDD456 pKa = 3.44SIITHH461 pKa = 6.8LPLGQEE467 pKa = 4.5FGCYY471 pKa = 8.44RR472 pKa = 11.84VGSVKK477 pKa = 9.1THH479 pKa = 5.11VQCKK483 pKa = 8.49PYY485 pKa = 10.28HH486 pKa = 5.96HH487 pKa = 7.11AFDD490 pKa = 4.58GEE492 pKa = 4.51KK493 pKa = 8.84TCNVSSDD500 pKa = 3.37GDD502 pKa = 3.78CRR504 pKa = 11.84EE505 pKa = 4.39GEE507 pKa = 3.82LCAAVKK513 pKa = 10.82LNGQGIITARR523 pKa = 11.84THH525 pKa = 5.76LGEE528 pKa = 4.27VQVKK532 pKa = 9.81HH533 pKa = 6.47CLEE536 pKa = 4.08EE537 pKa = 4.31CQFSFTRR544 pKa = 11.84VNDD547 pKa = 3.69LEE549 pKa = 4.63VSFTCPDD556 pKa = 3.15GQQRR560 pKa = 11.84RR561 pKa = 11.84LHH563 pKa = 6.29SNAVDD568 pKa = 3.88TNCPFQNKK576 pKa = 9.62LGVYY580 pKa = 9.9ALYY583 pKa = 10.49VCRR586 pKa = 11.84ATHH589 pKa = 6.56RR590 pKa = 11.84PLVLYY595 pKa = 7.41TTFIWFVVGTIVLATSLQMISVTVKK620 pKa = 9.1LYY622 pKa = 10.79CYY624 pKa = 10.31LVVLAKK630 pKa = 10.47RR631 pKa = 11.84KK632 pKa = 9.88LDD634 pKa = 3.51TGKK637 pKa = 10.69GVCTDD642 pKa = 3.48CGEE645 pKa = 4.61SVLSTEE651 pKa = 3.72EE652 pKa = 3.98WQRR655 pKa = 11.84HH656 pKa = 3.89QACKK660 pKa = 10.13KK661 pKa = 9.93GKK663 pKa = 9.93CPYY666 pKa = 10.0CGVKK670 pKa = 10.7GSGNDD675 pKa = 3.52VQKK678 pKa = 11.06HH679 pKa = 5.39VINCLQRR686 pKa = 11.84EE687 pKa = 4.44TVLEE691 pKa = 4.16HH692 pKa = 7.1DD693 pKa = 4.94LNVLTIRR700 pKa = 11.84RR701 pKa = 11.84TPRR704 pKa = 11.84YY705 pKa = 9.55ALRR708 pKa = 11.84LGCFLNALQGRR719 pKa = 11.84PIRR722 pKa = 11.84LIWLVVLLLLFVFLIKK738 pKa = 10.06PVRR741 pKa = 11.84SLEE744 pKa = 4.35TNDD747 pKa = 3.17QTEE750 pKa = 4.4GLWEE754 pKa = 3.97EE755 pKa = 4.77GIEE758 pKa = 4.05EE759 pKa = 4.28VEE761 pKa = 4.39KK762 pKa = 10.98CGKK765 pKa = 9.29GCWYY769 pKa = 10.25NQDD772 pKa = 2.76VCTCDD777 pKa = 3.53KK778 pKa = 10.99EE779 pKa = 5.0EE780 pKa = 4.29PLHH783 pKa = 5.4TSRR786 pKa = 11.84HH787 pKa = 5.15ILSVPAKK794 pKa = 10.35DD795 pKa = 3.75DD796 pKa = 3.8TEE798 pKa = 4.23KK799 pKa = 10.62ATSKK803 pKa = 10.56DD804 pKa = 3.54RR805 pKa = 11.84QGKK808 pKa = 9.44KK809 pKa = 8.0VMRR812 pKa = 11.84SLDD815 pKa = 3.51VEE817 pKa = 4.55APWGTLHH824 pKa = 7.41IPEE827 pKa = 4.6TFSPAGSVKK836 pKa = 10.22HH837 pKa = 6.57ISLSWEE843 pKa = 3.76SSRR846 pKa = 11.84VVGKK850 pKa = 10.33RR851 pKa = 11.84VILSGKK857 pKa = 8.1STAILKK863 pKa = 9.37LNPKK867 pKa = 9.24TSTSWEE873 pKa = 4.19MTSPDD878 pKa = 3.59ANEE881 pKa = 4.15KK882 pKa = 10.61KK883 pKa = 10.38ILTLSILDD891 pKa = 3.48YY892 pKa = 9.52TQIYY896 pKa = 9.59SSRR899 pKa = 11.84FEE901 pKa = 4.2YY902 pKa = 9.88LTGDD906 pKa = 3.48RR907 pKa = 11.84KK908 pKa = 9.28VTTWSEE914 pKa = 4.65GSCTGPCPKK923 pKa = 10.45DD924 pKa = 3.65CGCNDD929 pKa = 4.06PSCHH933 pKa = 5.82TKK935 pKa = 9.79QWLNTRR941 pKa = 11.84NWRR944 pKa = 11.84CNPTWCWGIGTGCSCCSAKK963 pKa = 10.55VVDD966 pKa = 5.93LYY968 pKa = 11.7KK969 pKa = 10.87NWLVSIWQIEE979 pKa = 4.35HH980 pKa = 6.57LRR982 pKa = 11.84TPVVACLEE990 pKa = 4.12FDD992 pKa = 3.63HH993 pKa = 6.81EE994 pKa = 4.36NRR996 pKa = 11.84VCDD999 pKa = 3.71VVEE1002 pKa = 4.37AGIEE1006 pKa = 4.06IQLGPVTVAFSDD1018 pKa = 3.62PFGEE1022 pKa = 4.8QKK1024 pKa = 10.41ILPQRR1029 pKa = 11.84IAVYY1033 pKa = 9.91HH1034 pKa = 6.14KK1035 pKa = 9.8RR1036 pKa = 11.84DD1037 pKa = 3.27VDD1039 pKa = 3.8HH1040 pKa = 6.68EE1041 pKa = 4.41HH1042 pKa = 6.97VDD1044 pKa = 3.61LLHH1047 pKa = 5.83NHH1049 pKa = 7.44GIGGAEE1055 pKa = 4.37QYY1057 pKa = 10.68CKK1059 pKa = 10.55LQSCTHH1065 pKa = 5.87GTAGDD1070 pKa = 3.8YY1071 pKa = 10.87QIINPDD1077 pKa = 3.41ALVFDD1082 pKa = 6.57DD1083 pKa = 4.28ITSMNYY1089 pKa = 9.2FKK1091 pKa = 11.17KK1092 pKa = 10.57LDD1094 pKa = 3.78VYY1096 pKa = 11.36NKK1098 pKa = 10.14LWMSWEE1104 pKa = 4.09GVNLGYY1110 pKa = 10.69YY1111 pKa = 9.61CNPGDD1116 pKa = 3.74WTTCTAEE1123 pKa = 4.19NIVVRR1128 pKa = 11.84NSEE1131 pKa = 4.01AFSNRR1136 pKa = 11.84NNLEE1140 pKa = 3.75RR1141 pKa = 11.84NYY1143 pKa = 10.41SVSHH1147 pKa = 6.7FFHH1150 pKa = 7.04SSRR1153 pKa = 11.84VYY1155 pKa = 10.64GSGKK1159 pKa = 10.02SLVMDD1164 pKa = 4.22LKK1166 pKa = 10.99GRR1168 pKa = 11.84PIQSGGNINVYY1179 pKa = 8.67VTVNNLEE1186 pKa = 4.29LNSKK1190 pKa = 10.11KK1191 pKa = 10.75VVITGLKK1198 pKa = 9.57VNLRR1202 pKa = 11.84ACTGCFGCNLGAEE1215 pKa = 4.58CQISLSLTEE1224 pKa = 4.29PDD1226 pKa = 4.02EE1227 pKa = 4.22FHH1229 pKa = 6.89LHH1231 pKa = 6.06LKK1233 pKa = 10.35SVTPGVTVPDD1243 pKa = 3.5TSFLVTSSEE1252 pKa = 3.59EE1253 pKa = 3.56KK1254 pKa = 9.77MFNIRR1259 pKa = 11.84VFSILKK1265 pKa = 9.92DD1266 pKa = 3.45VNFCIEE1272 pKa = 4.03VLEE1275 pKa = 4.73SKK1277 pKa = 10.31HH1278 pKa = 7.21CPEE1281 pKa = 5.16CDD1283 pKa = 3.27KK1284 pKa = 11.41KK1285 pKa = 10.97DD1286 pKa = 3.81LQSCLQLTFEE1296 pKa = 4.71DD1297 pKa = 4.38PKK1299 pKa = 11.0PVLLEE1304 pKa = 3.86HH1305 pKa = 7.01RR1306 pKa = 11.84SVLFSKK1312 pKa = 10.84SNQTCGDD1319 pKa = 3.61STISCWSSSAGILFKK1334 pKa = 11.2GIGNFLSTHH1343 pKa = 6.52FGSIFRR1349 pKa = 11.84GILLSVLPILLIVGLIFFSPQIISLMRR1376 pKa = 11.84LCKK1379 pKa = 9.99RR1380 pKa = 11.84GRR1382 pKa = 11.84SVVGFRR1388 pKa = 11.84KK1389 pKa = 9.92RR1390 pKa = 11.84FYY1392 pKa = 11.13KK1393 pKa = 10.55PLTDD1397 pKa = 3.9GADD1400 pKa = 3.1MGLSAEE1406 pKa = 4.39EE1407 pKa = 4.24KK1408 pKa = 10.82AFLTGIFGKK1417 pKa = 10.53KK1418 pKa = 9.5KK1419 pKa = 9.51EE1420 pKa = 4.07
MM1 pKa = 6.81EE2 pKa = 5.24TYY4 pKa = 10.29HH5 pKa = 6.68GSRR8 pKa = 11.84YY9 pKa = 7.43TATSCLFFVIMLVVAITADD28 pKa = 3.58NNVTEE33 pKa = 4.43TTTAEE38 pKa = 4.36VVSNTTGSTEE48 pKa = 4.16GDD50 pKa = 3.67TTSKK54 pKa = 11.15GMDD57 pKa = 3.27TTQIPQINQTSATTRR72 pKa = 11.84SNNITTKK79 pKa = 9.21TSRR82 pKa = 11.84SEE84 pKa = 4.18SGVDD88 pKa = 3.64PSATQPLLVSSQASGTIGQQRR109 pKa = 11.84GLSPTPDD116 pKa = 3.28PGARR120 pKa = 11.84IRR122 pKa = 11.84VKK124 pKa = 10.11GGKK127 pKa = 8.98RR128 pKa = 11.84RR129 pKa = 11.84AEE131 pKa = 3.81RR132 pKa = 11.84LTARR136 pKa = 11.84LDD138 pKa = 3.6VNGNRR143 pKa = 11.84VVLKK147 pKa = 10.78RR148 pKa = 11.84EE149 pKa = 3.59ISGNLQLAKK158 pKa = 10.22EE159 pKa = 4.05QLEE162 pKa = 4.18EE163 pKa = 3.97AVLNMEE169 pKa = 4.75IDD171 pKa = 3.86SLDD174 pKa = 3.73LEE176 pKa = 4.35QLEE179 pKa = 4.57EE180 pKa = 3.97EE181 pKa = 4.29SPYY184 pKa = 11.09RR185 pKa = 11.84SMQRR189 pKa = 11.84TRR191 pKa = 11.84LMLPKK196 pKa = 10.3LEE198 pKa = 4.03SFAYY202 pKa = 10.47NEE204 pKa = 4.07EE205 pKa = 4.03QEE207 pKa = 4.25EE208 pKa = 4.36DD209 pKa = 3.92VMVTGKK215 pKa = 10.62LSRR218 pKa = 11.84FTRR221 pKa = 11.84FEE223 pKa = 4.43RR224 pKa = 11.84INIDD228 pKa = 3.15KK229 pKa = 10.84LEE231 pKa = 4.79SITVFQNLTNRR242 pKa = 11.84SVLTLCTYY250 pKa = 10.47FGHH253 pKa = 6.63YY254 pKa = 9.23YY255 pKa = 10.93NKK257 pKa = 8.65TLKK260 pKa = 9.1GTTVSIHH267 pKa = 6.1HH268 pKa = 6.64KK269 pKa = 10.52CDD271 pKa = 3.71IEE273 pKa = 4.48TQCKK277 pKa = 10.05GLTEE281 pKa = 4.23HH282 pKa = 6.75VSIDD286 pKa = 4.48RR287 pKa = 11.84ITNYY291 pKa = 11.17DD292 pKa = 3.6NIKK295 pKa = 9.9IINIMEE301 pKa = 4.29LPVIIIAYY309 pKa = 5.52FTRR312 pKa = 11.84HH313 pKa = 6.03FGFTYY318 pKa = 10.11KK319 pKa = 10.71NLVIHH324 pKa = 6.14EE325 pKa = 4.31QIEE328 pKa = 4.41NCLFFYY334 pKa = 9.82PQNPVACMEE343 pKa = 4.41NNWGGRR349 pKa = 11.84SHH351 pKa = 7.13PVVHH355 pKa = 6.57LVIEE359 pKa = 4.62RR360 pKa = 11.84KK361 pKa = 9.12YY362 pKa = 11.01LKK364 pKa = 10.07ADD366 pKa = 3.55SSITLCGLKK375 pKa = 10.18KK376 pKa = 10.78GNPTSLKK383 pKa = 9.01GTWFAEE389 pKa = 4.3KK390 pKa = 10.06TKK392 pKa = 10.04VTIDD396 pKa = 3.12LGEE399 pKa = 4.21PQTAGRR405 pKa = 11.84RR406 pKa = 11.84KK407 pKa = 9.71LLAIEE412 pKa = 4.09RR413 pKa = 11.84RR414 pKa = 11.84RR415 pKa = 11.84RR416 pKa = 11.84LPGRR420 pKa = 11.84EE421 pKa = 4.51DD422 pKa = 3.21IRR424 pKa = 11.84CHH426 pKa = 6.73SGSHH430 pKa = 5.66LVKK433 pKa = 9.49IDD435 pKa = 3.31KK436 pKa = 9.64YY437 pKa = 11.14APTNEE442 pKa = 4.08FQSFPNAKK450 pKa = 9.55IGFCNDD456 pKa = 3.44SIITHH461 pKa = 6.8LPLGQEE467 pKa = 4.5FGCYY471 pKa = 8.44RR472 pKa = 11.84VGSVKK477 pKa = 9.1THH479 pKa = 5.11VQCKK483 pKa = 8.49PYY485 pKa = 10.28HH486 pKa = 5.96HH487 pKa = 7.11AFDD490 pKa = 4.58GEE492 pKa = 4.51KK493 pKa = 8.84TCNVSSDD500 pKa = 3.37GDD502 pKa = 3.78CRR504 pKa = 11.84EE505 pKa = 4.39GEE507 pKa = 3.82LCAAVKK513 pKa = 10.82LNGQGIITARR523 pKa = 11.84THH525 pKa = 5.76LGEE528 pKa = 4.27VQVKK532 pKa = 9.81HH533 pKa = 6.47CLEE536 pKa = 4.08EE537 pKa = 4.31CQFSFTRR544 pKa = 11.84VNDD547 pKa = 3.69LEE549 pKa = 4.63VSFTCPDD556 pKa = 3.15GQQRR560 pKa = 11.84RR561 pKa = 11.84LHH563 pKa = 6.29SNAVDD568 pKa = 3.88TNCPFQNKK576 pKa = 9.62LGVYY580 pKa = 9.9ALYY583 pKa = 10.49VCRR586 pKa = 11.84ATHH589 pKa = 6.56RR590 pKa = 11.84PLVLYY595 pKa = 7.41TTFIWFVVGTIVLATSLQMISVTVKK620 pKa = 9.1LYY622 pKa = 10.79CYY624 pKa = 10.31LVVLAKK630 pKa = 10.47RR631 pKa = 11.84KK632 pKa = 9.88LDD634 pKa = 3.51TGKK637 pKa = 10.69GVCTDD642 pKa = 3.48CGEE645 pKa = 4.61SVLSTEE651 pKa = 3.72EE652 pKa = 3.98WQRR655 pKa = 11.84HH656 pKa = 3.89QACKK660 pKa = 10.13KK661 pKa = 9.93GKK663 pKa = 9.93CPYY666 pKa = 10.0CGVKK670 pKa = 10.7GSGNDD675 pKa = 3.52VQKK678 pKa = 11.06HH679 pKa = 5.39VINCLQRR686 pKa = 11.84EE687 pKa = 4.44TVLEE691 pKa = 4.16HH692 pKa = 7.1DD693 pKa = 4.94LNVLTIRR700 pKa = 11.84RR701 pKa = 11.84TPRR704 pKa = 11.84YY705 pKa = 9.55ALRR708 pKa = 11.84LGCFLNALQGRR719 pKa = 11.84PIRR722 pKa = 11.84LIWLVVLLLLFVFLIKK738 pKa = 10.06PVRR741 pKa = 11.84SLEE744 pKa = 4.35TNDD747 pKa = 3.17QTEE750 pKa = 4.4GLWEE754 pKa = 3.97EE755 pKa = 4.77GIEE758 pKa = 4.05EE759 pKa = 4.28VEE761 pKa = 4.39KK762 pKa = 10.98CGKK765 pKa = 9.29GCWYY769 pKa = 10.25NQDD772 pKa = 2.76VCTCDD777 pKa = 3.53KK778 pKa = 10.99EE779 pKa = 5.0EE780 pKa = 4.29PLHH783 pKa = 5.4TSRR786 pKa = 11.84HH787 pKa = 5.15ILSVPAKK794 pKa = 10.35DD795 pKa = 3.75DD796 pKa = 3.8TEE798 pKa = 4.23KK799 pKa = 10.62ATSKK803 pKa = 10.56DD804 pKa = 3.54RR805 pKa = 11.84QGKK808 pKa = 9.44KK809 pKa = 8.0VMRR812 pKa = 11.84SLDD815 pKa = 3.51VEE817 pKa = 4.55APWGTLHH824 pKa = 7.41IPEE827 pKa = 4.6TFSPAGSVKK836 pKa = 10.22HH837 pKa = 6.57ISLSWEE843 pKa = 3.76SSRR846 pKa = 11.84VVGKK850 pKa = 10.33RR851 pKa = 11.84VILSGKK857 pKa = 8.1STAILKK863 pKa = 9.37LNPKK867 pKa = 9.24TSTSWEE873 pKa = 4.19MTSPDD878 pKa = 3.59ANEE881 pKa = 4.15KK882 pKa = 10.61KK883 pKa = 10.38ILTLSILDD891 pKa = 3.48YY892 pKa = 9.52TQIYY896 pKa = 9.59SSRR899 pKa = 11.84FEE901 pKa = 4.2YY902 pKa = 9.88LTGDD906 pKa = 3.48RR907 pKa = 11.84KK908 pKa = 9.28VTTWSEE914 pKa = 4.65GSCTGPCPKK923 pKa = 10.45DD924 pKa = 3.65CGCNDD929 pKa = 4.06PSCHH933 pKa = 5.82TKK935 pKa = 9.79QWLNTRR941 pKa = 11.84NWRR944 pKa = 11.84CNPTWCWGIGTGCSCCSAKK963 pKa = 10.55VVDD966 pKa = 5.93LYY968 pKa = 11.7KK969 pKa = 10.87NWLVSIWQIEE979 pKa = 4.35HH980 pKa = 6.57LRR982 pKa = 11.84TPVVACLEE990 pKa = 4.12FDD992 pKa = 3.63HH993 pKa = 6.81EE994 pKa = 4.36NRR996 pKa = 11.84VCDD999 pKa = 3.71VVEE1002 pKa = 4.37AGIEE1006 pKa = 4.06IQLGPVTVAFSDD1018 pKa = 3.62PFGEE1022 pKa = 4.8QKK1024 pKa = 10.41ILPQRR1029 pKa = 11.84IAVYY1033 pKa = 9.91HH1034 pKa = 6.14KK1035 pKa = 9.8RR1036 pKa = 11.84DD1037 pKa = 3.27VDD1039 pKa = 3.8HH1040 pKa = 6.68EE1041 pKa = 4.41HH1042 pKa = 6.97VDD1044 pKa = 3.61LLHH1047 pKa = 5.83NHH1049 pKa = 7.44GIGGAEE1055 pKa = 4.37QYY1057 pKa = 10.68CKK1059 pKa = 10.55LQSCTHH1065 pKa = 5.87GTAGDD1070 pKa = 3.8YY1071 pKa = 10.87QIINPDD1077 pKa = 3.41ALVFDD1082 pKa = 6.57DD1083 pKa = 4.28ITSMNYY1089 pKa = 9.2FKK1091 pKa = 11.17KK1092 pKa = 10.57LDD1094 pKa = 3.78VYY1096 pKa = 11.36NKK1098 pKa = 10.14LWMSWEE1104 pKa = 4.09GVNLGYY1110 pKa = 10.69YY1111 pKa = 9.61CNPGDD1116 pKa = 3.74WTTCTAEE1123 pKa = 4.19NIVVRR1128 pKa = 11.84NSEE1131 pKa = 4.01AFSNRR1136 pKa = 11.84NNLEE1140 pKa = 3.75RR1141 pKa = 11.84NYY1143 pKa = 10.41SVSHH1147 pKa = 6.7FFHH1150 pKa = 7.04SSRR1153 pKa = 11.84VYY1155 pKa = 10.64GSGKK1159 pKa = 10.02SLVMDD1164 pKa = 4.22LKK1166 pKa = 10.99GRR1168 pKa = 11.84PIQSGGNINVYY1179 pKa = 8.67VTVNNLEE1186 pKa = 4.29LNSKK1190 pKa = 10.11KK1191 pKa = 10.75VVITGLKK1198 pKa = 9.57VNLRR1202 pKa = 11.84ACTGCFGCNLGAEE1215 pKa = 4.58CQISLSLTEE1224 pKa = 4.29PDD1226 pKa = 4.02EE1227 pKa = 4.22FHH1229 pKa = 6.89LHH1231 pKa = 6.06LKK1233 pKa = 10.35SVTPGVTVPDD1243 pKa = 3.5TSFLVTSSEE1252 pKa = 3.59EE1253 pKa = 3.56KK1254 pKa = 9.77MFNIRR1259 pKa = 11.84VFSILKK1265 pKa = 9.92DD1266 pKa = 3.45VNFCIEE1272 pKa = 4.03VLEE1275 pKa = 4.73SKK1277 pKa = 10.31HH1278 pKa = 7.21CPEE1281 pKa = 5.16CDD1283 pKa = 3.27KK1284 pKa = 11.41KK1285 pKa = 10.97DD1286 pKa = 3.81LQSCLQLTFEE1296 pKa = 4.71DD1297 pKa = 4.38PKK1299 pKa = 11.0PVLLEE1304 pKa = 3.86HH1305 pKa = 7.01RR1306 pKa = 11.84SVLFSKK1312 pKa = 10.84SNQTCGDD1319 pKa = 3.61STISCWSSSAGILFKK1334 pKa = 11.2GIGNFLSTHH1343 pKa = 6.52FGSIFRR1349 pKa = 11.84GILLSVLPILLIVGLIFFSPQIISLMRR1376 pKa = 11.84LCKK1379 pKa = 9.99RR1380 pKa = 11.84GRR1382 pKa = 11.84SVVGFRR1388 pKa = 11.84KK1389 pKa = 9.92RR1390 pKa = 11.84FYY1392 pKa = 11.13KK1393 pKa = 10.55PLTDD1397 pKa = 3.9GADD1400 pKa = 3.1MGLSAEE1406 pKa = 4.39EE1407 pKa = 4.24KK1408 pKa = 10.82AFLTGIFGKK1417 pKa = 10.53KK1418 pKa = 9.5KK1419 pKa = 9.51EE1420 pKa = 4.07
Molecular weight: 159.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5901 |
514 |
3967 |
1967.0 |
222.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.118 ± 1.254 | 2.61 ± 0.596 |
5.05 ± 0.309 | 7.202 ± 0.367 |
4.203 ± 0.135 | 5.287 ± 0.895 |
2.135 ± 0.359 | 5.965 ± 0.262 |
7.541 ± 0.519 | 10.185 ± 1.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.339 ± 0.433 | 5.118 ± 0.479 |
3.474 ± 0.224 | 3.474 ± 0.024 |
4.931 ± 0.408 | 8.863 ± 0.678 |
5.626 ± 0.878 | 6.779 ± 0.44 |
1.288 ± 0.364 | 2.813 ± 0.109 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
