
Only Syngen Nebraska virus 5
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Algavirales; Phycodnaviridae; Chlorovirus; unclassified Chlorovirus
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
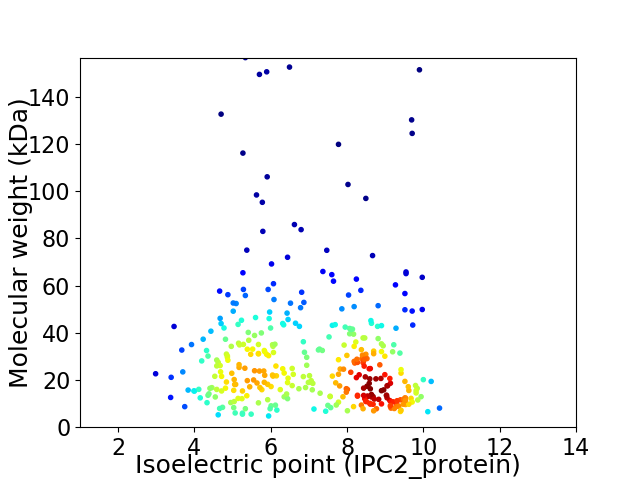
Virtual 2D-PAGE plot for 355 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J0F9M0|A0A1J0F9M0_9PHYC Uncharacterized protein OS=Only Syngen Nebraska virus 5 OX=1917232 GN=OS5_176L PE=4 SV=1
MM1 pKa = 7.68GLPPLEE7 pKa = 4.69LVLNDD12 pKa = 3.98DD13 pKa = 3.65EE14 pKa = 4.99RR15 pKa = 11.84ALGVLYY21 pKa = 10.59EE22 pKa = 4.31GALTGEE28 pKa = 4.73ALLALPGVLYY38 pKa = 10.85DD39 pKa = 4.94GVLPLPGVLTGEE51 pKa = 4.68GEE53 pKa = 4.43LDD55 pKa = 3.58RR56 pKa = 11.84PLAPLLLLPDD66 pKa = 4.87PFQEE70 pKa = 4.63TILLDD75 pKa = 3.54IYY77 pKa = 10.09YY78 pKa = 10.07IKK80 pKa = 10.5II81 pKa = 3.56
MM1 pKa = 7.68GLPPLEE7 pKa = 4.69LVLNDD12 pKa = 3.98DD13 pKa = 3.65EE14 pKa = 4.99RR15 pKa = 11.84ALGVLYY21 pKa = 10.59EE22 pKa = 4.31GALTGEE28 pKa = 4.73ALLALPGVLYY38 pKa = 10.85DD39 pKa = 4.94GVLPLPGVLTGEE51 pKa = 4.68GEE53 pKa = 4.43LDD55 pKa = 3.58RR56 pKa = 11.84PLAPLLLLPDD66 pKa = 4.87PFQEE70 pKa = 4.63TILLDD75 pKa = 3.54IYY77 pKa = 10.09YY78 pKa = 10.07IKK80 pKa = 10.5II81 pKa = 3.56
Molecular weight: 8.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J0FA23|A0A1J0FA23_9PHYC Uncharacterized protein OS=Only Syngen Nebraska virus 5 OX=1917232 GN=OS5_322L PE=4 SV=1
MM1 pKa = 7.38DD2 pKa = 3.84TKK4 pKa = 10.88LRR6 pKa = 11.84RR7 pKa = 11.84QTLKK11 pKa = 10.09RR12 pKa = 11.84DD13 pKa = 3.29EE14 pKa = 4.81RR15 pKa = 11.84KK16 pKa = 10.06LSTRR20 pKa = 11.84EE21 pKa = 3.71RR22 pKa = 11.84QEE24 pKa = 4.32LEE26 pKa = 3.86RR27 pKa = 11.84QRR29 pKa = 11.84LTRR32 pKa = 11.84MRR34 pKa = 11.84RR35 pKa = 11.84DD36 pKa = 3.75AKK38 pKa = 10.08PMPMIINSKK47 pKa = 6.99TPRR50 pKa = 11.84TVAPVRR56 pKa = 11.84PRR58 pKa = 11.84SRR60 pKa = 11.84SPPSLFKK67 pKa = 10.72RR68 pKa = 11.84IGSGGIKK75 pKa = 10.16KK76 pKa = 9.54IRR78 pKa = 11.84QQTIKK83 pKa = 10.61NEE85 pKa = 4.02RR86 pKa = 11.84GRR88 pKa = 11.84TYY90 pKa = 10.88AVKK93 pKa = 10.4KK94 pKa = 8.35ITGPRR99 pKa = 11.84VSCSKK104 pKa = 10.9VLTFAQQGAICWFTALITTLFFSQYY129 pKa = 8.25TRR131 pKa = 11.84VVMKK135 pKa = 9.44IHH137 pKa = 6.37ARR139 pKa = 11.84QMVKK143 pKa = 10.53YY144 pKa = 10.33SNTKK148 pKa = 10.05QIAEE152 pKa = 5.05AILEE156 pKa = 4.55LIKK159 pKa = 10.93GYY161 pKa = 10.93DD162 pKa = 3.15SGKK165 pKa = 8.2VSRR168 pKa = 11.84RR169 pKa = 11.84VVEE172 pKa = 4.33SMQPHH177 pKa = 5.91QFLKK181 pKa = 10.64DD182 pKa = 3.4LRR184 pKa = 11.84ATRR187 pKa = 11.84SNLFDD192 pKa = 3.3STLNGSSEE200 pKa = 3.95AHH202 pKa = 5.72YY203 pKa = 10.83NPYY206 pKa = 8.62VHH208 pKa = 7.36KK209 pKa = 10.41ILGFLRR215 pKa = 11.84VPHH218 pKa = 6.88LGLGIVGGKK227 pKa = 9.03IVYY230 pKa = 10.41SGFNVDD236 pKa = 5.49LPLDD240 pKa = 4.05SKK242 pKa = 11.32LWAQAMKK249 pKa = 8.77TQSPRR254 pKa = 11.84GTFVDD259 pKa = 3.83TNNPEE264 pKa = 3.81VLLLHH269 pKa = 7.03RR270 pKa = 11.84DD271 pKa = 3.3GGEE274 pKa = 4.01EE275 pKa = 4.06YY276 pKa = 11.16VQGLWGRR283 pKa = 11.84PRR285 pKa = 11.84PAIGSVAGISSDD297 pKa = 2.81IHH299 pKa = 6.79NKK301 pKa = 8.97FIKK304 pKa = 10.59YY305 pKa = 9.9NGRR308 pKa = 11.84NYY310 pKa = 10.24ILDD313 pKa = 3.58SCILGGEE320 pKa = 4.26VRR322 pKa = 11.84TPSCSVAHH330 pKa = 7.16AIAGVTCNNKK340 pKa = 9.49RR341 pKa = 11.84FIYY344 pKa = 10.29NGWTARR350 pKa = 11.84SADD353 pKa = 3.61PAMSGSGSVIRR364 pKa = 11.84DD365 pKa = 3.77MPCSLMPADD374 pKa = 3.42WASNKK379 pKa = 9.67SFCINTSACKK389 pKa = 10.15MNIAKK394 pKa = 9.89PNQLSKK400 pKa = 10.41EE401 pKa = 4.06LCFNGVARR409 pKa = 11.84STVVYY414 pKa = 10.23IRR416 pKa = 11.84SDD418 pKa = 3.43LAKK421 pKa = 10.41LGGYY425 pKa = 9.25KK426 pKa = 10.46VIGKK430 pKa = 9.17QIQRR434 pKa = 11.84VRR436 pKa = 11.84KK437 pKa = 8.83RR438 pKa = 3.17
MM1 pKa = 7.38DD2 pKa = 3.84TKK4 pKa = 10.88LRR6 pKa = 11.84RR7 pKa = 11.84QTLKK11 pKa = 10.09RR12 pKa = 11.84DD13 pKa = 3.29EE14 pKa = 4.81RR15 pKa = 11.84KK16 pKa = 10.06LSTRR20 pKa = 11.84EE21 pKa = 3.71RR22 pKa = 11.84QEE24 pKa = 4.32LEE26 pKa = 3.86RR27 pKa = 11.84QRR29 pKa = 11.84LTRR32 pKa = 11.84MRR34 pKa = 11.84RR35 pKa = 11.84DD36 pKa = 3.75AKK38 pKa = 10.08PMPMIINSKK47 pKa = 6.99TPRR50 pKa = 11.84TVAPVRR56 pKa = 11.84PRR58 pKa = 11.84SRR60 pKa = 11.84SPPSLFKK67 pKa = 10.72RR68 pKa = 11.84IGSGGIKK75 pKa = 10.16KK76 pKa = 9.54IRR78 pKa = 11.84QQTIKK83 pKa = 10.61NEE85 pKa = 4.02RR86 pKa = 11.84GRR88 pKa = 11.84TYY90 pKa = 10.88AVKK93 pKa = 10.4KK94 pKa = 8.35ITGPRR99 pKa = 11.84VSCSKK104 pKa = 10.9VLTFAQQGAICWFTALITTLFFSQYY129 pKa = 8.25TRR131 pKa = 11.84VVMKK135 pKa = 9.44IHH137 pKa = 6.37ARR139 pKa = 11.84QMVKK143 pKa = 10.53YY144 pKa = 10.33SNTKK148 pKa = 10.05QIAEE152 pKa = 5.05AILEE156 pKa = 4.55LIKK159 pKa = 10.93GYY161 pKa = 10.93DD162 pKa = 3.15SGKK165 pKa = 8.2VSRR168 pKa = 11.84RR169 pKa = 11.84VVEE172 pKa = 4.33SMQPHH177 pKa = 5.91QFLKK181 pKa = 10.64DD182 pKa = 3.4LRR184 pKa = 11.84ATRR187 pKa = 11.84SNLFDD192 pKa = 3.3STLNGSSEE200 pKa = 3.95AHH202 pKa = 5.72YY203 pKa = 10.83NPYY206 pKa = 8.62VHH208 pKa = 7.36KK209 pKa = 10.41ILGFLRR215 pKa = 11.84VPHH218 pKa = 6.88LGLGIVGGKK227 pKa = 9.03IVYY230 pKa = 10.41SGFNVDD236 pKa = 5.49LPLDD240 pKa = 4.05SKK242 pKa = 11.32LWAQAMKK249 pKa = 8.77TQSPRR254 pKa = 11.84GTFVDD259 pKa = 3.83TNNPEE264 pKa = 3.81VLLLHH269 pKa = 7.03RR270 pKa = 11.84DD271 pKa = 3.3GGEE274 pKa = 4.01EE275 pKa = 4.06YY276 pKa = 11.16VQGLWGRR283 pKa = 11.84PRR285 pKa = 11.84PAIGSVAGISSDD297 pKa = 2.81IHH299 pKa = 6.79NKK301 pKa = 8.97FIKK304 pKa = 10.59YY305 pKa = 9.9NGRR308 pKa = 11.84NYY310 pKa = 10.24ILDD313 pKa = 3.58SCILGGEE320 pKa = 4.26VRR322 pKa = 11.84TPSCSVAHH330 pKa = 7.16AIAGVTCNNKK340 pKa = 9.49RR341 pKa = 11.84FIYY344 pKa = 10.29NGWTARR350 pKa = 11.84SADD353 pKa = 3.61PAMSGSGSVIRR364 pKa = 11.84DD365 pKa = 3.77MPCSLMPADD374 pKa = 3.42WASNKK379 pKa = 9.67SFCINTSACKK389 pKa = 10.15MNIAKK394 pKa = 9.89PNQLSKK400 pKa = 10.41EE401 pKa = 4.06LCFNGVARR409 pKa = 11.84STVVYY414 pKa = 10.23IRR416 pKa = 11.84SDD418 pKa = 3.43LAKK421 pKa = 10.41LGGYY425 pKa = 9.25KK426 pKa = 10.46VIGKK430 pKa = 9.17QIQRR434 pKa = 11.84VRR436 pKa = 11.84KK437 pKa = 8.83RR438 pKa = 3.17
Molecular weight: 49.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
98420 |
42 |
1506 |
277.2 |
31.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.417 ± 0.153 | 1.43 ± 0.09 |
5.712 ± 0.182 | 5.439 ± 0.233 |
4.583 ± 0.1 | 6.514 ± 0.26 |
1.736 ± 0.087 | 6.973 ± 0.128 |
7.901 ± 0.332 | 7.064 ± 0.119 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.595 ± 0.078 | 6.014 ± 0.258 |
5.141 ± 0.237 | 2.878 ± 0.097 |
4.201 ± 0.129 | 6.842 ± 0.147 |
6.527 ± 0.188 | 7.101 ± 0.149 |
1.071 ± 0.046 | 3.859 ± 0.101 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
