
Wenzhou narna-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.41
Get precalculated fractions of proteins
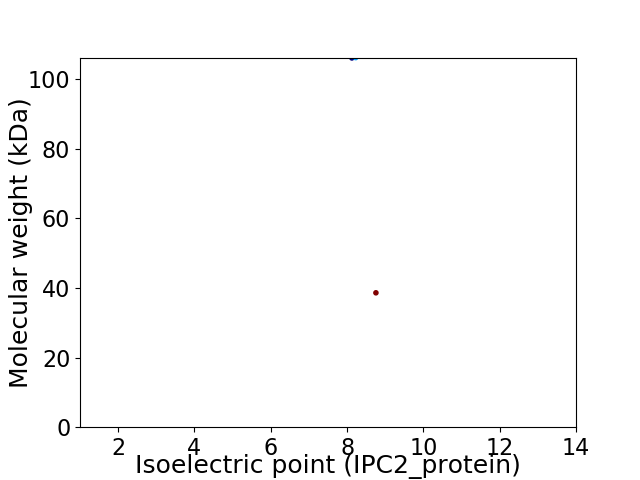
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJ26|A0A1L3KJ26_9VIRU Uncharacterized protein OS=Wenzhou narna-like virus 4 OX=1923579 PE=4 SV=1
MM1 pKa = 7.76QEE3 pKa = 4.17EE4 pKa = 5.25LSPQQNALQFLNRR17 pKa = 11.84ISAQSTIGMWTDD29 pKa = 3.09STPPFPCPSGKK40 pKa = 9.94KK41 pKa = 8.79EE42 pKa = 3.68GRR44 pKa = 11.84LAFVANTKK52 pKa = 10.58SGFNWLVKK60 pKa = 10.31LPKK63 pKa = 10.18SLLEE67 pKa = 4.1SVALEE72 pKa = 3.99RR73 pKa = 11.84VTTKK77 pKa = 10.65LGEE80 pKa = 4.04VRR82 pKa = 11.84TWQPQASEE90 pKa = 4.51SEE92 pKa = 4.54DD93 pKa = 3.84GIKK96 pKa = 8.94PTGLNYY102 pKa = 10.75SKK104 pKa = 10.95VILVPTIGDD113 pKa = 3.83CEE115 pKa = 4.32SWRR118 pKa = 11.84AAADD122 pKa = 3.99FCCGYY127 pKa = 10.65LSVIEE132 pKa = 4.63DD133 pKa = 3.96SYY135 pKa = 12.22AKK137 pKa = 10.24ARR139 pKa = 11.84KK140 pKa = 9.3LDD142 pKa = 3.57FHH144 pKa = 6.56TATTPFRR151 pKa = 11.84HH152 pKa = 5.78LVKK155 pKa = 9.63IDD157 pKa = 4.06KK158 pKa = 10.1SQQTAWIKK166 pKa = 10.8YY167 pKa = 5.6YY168 pKa = 9.85TAAPMSQILGEE179 pKa = 4.39TLPDD183 pKa = 3.71TPDD186 pKa = 3.04SSRR189 pKa = 11.84TPVFFSGKK197 pKa = 8.19FRR199 pKa = 11.84KK200 pKa = 9.37FMHH203 pKa = 5.21QRR205 pKa = 11.84CLKK208 pKa = 8.54GTRR211 pKa = 11.84DD212 pKa = 3.36LRR214 pKa = 11.84NIRR217 pKa = 11.84NAFNLLQGVKK227 pKa = 9.84KK228 pKa = 10.48ACQEE232 pKa = 3.62ISKK235 pKa = 10.74DD236 pKa = 4.14FILEE240 pKa = 3.98AYY242 pKa = 8.46KK243 pKa = 10.45KK244 pKa = 9.62HH245 pKa = 6.48RR246 pKa = 11.84KK247 pKa = 7.23TMEE250 pKa = 4.22TPCAGPPATEE260 pKa = 4.08LHH262 pKa = 4.77QAKK265 pKa = 10.5YY266 pKa = 10.68KK267 pKa = 9.76MLWTQGRR274 pKa = 11.84WIDD277 pKa = 2.99RR278 pKa = 11.84WKK280 pKa = 10.48KK281 pKa = 9.05YY282 pKa = 10.67KK283 pKa = 10.04PVLFKK288 pKa = 10.97PPGPAATLEE297 pKa = 4.41CPRR300 pKa = 11.84SEE302 pKa = 4.53GGGSQYY308 pKa = 11.32AFEE311 pKa = 4.2QNRR314 pKa = 11.84VFDD317 pKa = 3.8ARR319 pKa = 11.84EE320 pKa = 3.62IAVQSAIVNEE330 pKa = 4.04QLWFFFDD337 pKa = 3.41RR338 pKa = 11.84LGIRR342 pKa = 11.84DD343 pKa = 4.44AFLKK347 pKa = 7.73TQKK350 pKa = 9.65IDD352 pKa = 3.47EE353 pKa = 4.47VSPFWTWPEE362 pKa = 3.87SEE364 pKa = 4.07EE365 pKa = 4.09TGRR368 pKa = 11.84VQTVPTRR375 pKa = 11.84CLSPEE380 pKa = 3.85GKK382 pKa = 9.45IFYY385 pKa = 9.24QWQHH389 pKa = 5.73YY390 pKa = 9.18SPTWEE395 pKa = 4.18EE396 pKa = 4.33VFALYY401 pKa = 10.7NEE403 pKa = 4.44EE404 pKa = 4.14DD405 pKa = 3.69PKK407 pKa = 11.38NRR409 pKa = 11.84TVMVEE414 pKa = 4.18GIIEE418 pKa = 4.07PLKK421 pKa = 10.91LRR423 pKa = 11.84TITKK427 pKa = 9.54GPCHH431 pKa = 6.68RR432 pKa = 11.84KK433 pKa = 8.64WLSQSLQRR441 pKa = 11.84EE442 pKa = 4.17MADD445 pKa = 3.59CLDD448 pKa = 4.28GFWQFKK454 pKa = 10.0LNKK457 pKa = 9.79ANTDD461 pKa = 2.79HH462 pKa = 7.7RR463 pKa = 11.84MVTRR467 pKa = 11.84LNAACVRR474 pKa = 11.84LHH476 pKa = 6.54SKK478 pKa = 10.2QDD480 pKa = 3.05SFTPDD485 pKa = 2.9TPMWWCSGDD494 pKa = 3.69YY495 pKa = 10.59KK496 pKa = 11.0SATDD500 pKa = 4.2SISIHH505 pKa = 5.17HH506 pKa = 6.59TKK508 pKa = 10.6AALEE512 pKa = 4.07TLLNCTSPEE521 pKa = 4.05RR522 pKa = 11.84VNEE525 pKa = 3.92ACKK528 pKa = 10.25RR529 pKa = 11.84LYY531 pKa = 10.3RR532 pKa = 11.84DD533 pKa = 3.34EE534 pKa = 5.64LYY536 pKa = 10.64EE537 pKa = 5.24QIVQYY542 pKa = 9.52PKK544 pKa = 8.46WTGIPDD550 pKa = 3.51VQQVNGQLMGSVLSFPILCVINFVAYY576 pKa = 7.3WQSLEE581 pKa = 3.87EE582 pKa = 4.81HH583 pKa = 6.48YY584 pKa = 11.34GCTLNVRR591 pKa = 11.84SVPCLIHH598 pKa = 7.67GDD600 pKa = 4.08DD601 pKa = 3.9ILFKK605 pKa = 8.91TTHH608 pKa = 4.69EE609 pKa = 4.78HH610 pKa = 5.12YY611 pKa = 10.74AVWEE615 pKa = 4.07EE616 pKa = 4.12TIKK619 pKa = 11.16LFGLQKK625 pKa = 10.69SVGKK629 pKa = 10.4NYY631 pKa = 8.62FHH633 pKa = 7.28PKK635 pKa = 9.73VFTIDD640 pKa = 3.21SEE642 pKa = 4.47LWVEE646 pKa = 4.54SKK648 pKa = 10.81RR649 pKa = 11.84SDD651 pKa = 3.08GFHH654 pKa = 6.19FKK656 pKa = 10.24KK657 pKa = 10.86YY658 pKa = 10.44SGINVGSLLKK668 pKa = 10.84SKK670 pKa = 10.95VDD672 pKa = 3.49GRR674 pKa = 11.84NDD676 pKa = 3.54YY677 pKa = 10.68QRR679 pKa = 11.84LPIWDD684 pKa = 4.34KK685 pKa = 10.64FNSSIRR691 pKa = 11.84GAQDD695 pKa = 2.67KK696 pKa = 10.35EE697 pKa = 4.39LYY699 pKa = 10.12LKK701 pKa = 10.78RR702 pKa = 11.84FLHH705 pKa = 4.97YY706 pKa = 9.96HH707 pKa = 6.47KK708 pKa = 10.49STIKK712 pKa = 10.56PMTWTKK718 pKa = 11.15AGVLNLFLPRR728 pKa = 11.84EE729 pKa = 3.86RR730 pKa = 11.84GGIGFEE736 pKa = 4.82LPWKK740 pKa = 9.79PNEE743 pKa = 4.11IPIKK747 pKa = 10.54EE748 pKa = 4.27NGTPLVAVTKK758 pKa = 10.13HH759 pKa = 4.84QLNLASALCNSFRR772 pKa = 11.84EE773 pKa = 4.29AGPLKK778 pKa = 10.5SLAIVAVVDD787 pKa = 3.52KK788 pKa = 11.07TDD790 pKa = 2.91ATTEE794 pKa = 3.54KK795 pKa = 10.35RR796 pKa = 11.84YY797 pKa = 9.88RR798 pKa = 11.84LPFQEE803 pKa = 4.23VWKK806 pKa = 10.79HH807 pKa = 4.22EE808 pKa = 4.07STYY811 pKa = 11.09ARR813 pKa = 11.84EE814 pKa = 4.35IPGEE818 pKa = 3.96LADD821 pKa = 5.36PILSEE826 pKa = 4.37VPSGKK831 pKa = 9.47PEE833 pKa = 3.85EE834 pKa = 4.13YY835 pKa = 10.09RR836 pKa = 11.84LRR838 pKa = 11.84KK839 pKa = 9.62PEE841 pKa = 3.68IRR843 pKa = 11.84SLARR847 pKa = 11.84GPKK850 pKa = 8.81RR851 pKa = 11.84FRR853 pKa = 11.84VCTHH857 pKa = 6.73PPIAPNPRR865 pKa = 11.84IGRR868 pKa = 11.84IQRR871 pKa = 11.84PGEE874 pKa = 3.82LVVEE878 pKa = 4.67DD879 pKa = 4.58EE880 pKa = 4.93CVHH883 pKa = 6.58TYY885 pKa = 7.75PTEE888 pKa = 4.07IFSFPYY894 pKa = 9.75VRR896 pKa = 11.84SRR898 pKa = 11.84TIDD901 pKa = 3.32PDD903 pKa = 3.98VFWEE907 pKa = 4.58SIAWAANQYY916 pKa = 11.11NSGCSSTHH924 pKa = 5.18
MM1 pKa = 7.76QEE3 pKa = 4.17EE4 pKa = 5.25LSPQQNALQFLNRR17 pKa = 11.84ISAQSTIGMWTDD29 pKa = 3.09STPPFPCPSGKK40 pKa = 9.94KK41 pKa = 8.79EE42 pKa = 3.68GRR44 pKa = 11.84LAFVANTKK52 pKa = 10.58SGFNWLVKK60 pKa = 10.31LPKK63 pKa = 10.18SLLEE67 pKa = 4.1SVALEE72 pKa = 3.99RR73 pKa = 11.84VTTKK77 pKa = 10.65LGEE80 pKa = 4.04VRR82 pKa = 11.84TWQPQASEE90 pKa = 4.51SEE92 pKa = 4.54DD93 pKa = 3.84GIKK96 pKa = 8.94PTGLNYY102 pKa = 10.75SKK104 pKa = 10.95VILVPTIGDD113 pKa = 3.83CEE115 pKa = 4.32SWRR118 pKa = 11.84AAADD122 pKa = 3.99FCCGYY127 pKa = 10.65LSVIEE132 pKa = 4.63DD133 pKa = 3.96SYY135 pKa = 12.22AKK137 pKa = 10.24ARR139 pKa = 11.84KK140 pKa = 9.3LDD142 pKa = 3.57FHH144 pKa = 6.56TATTPFRR151 pKa = 11.84HH152 pKa = 5.78LVKK155 pKa = 9.63IDD157 pKa = 4.06KK158 pKa = 10.1SQQTAWIKK166 pKa = 10.8YY167 pKa = 5.6YY168 pKa = 9.85TAAPMSQILGEE179 pKa = 4.39TLPDD183 pKa = 3.71TPDD186 pKa = 3.04SSRR189 pKa = 11.84TPVFFSGKK197 pKa = 8.19FRR199 pKa = 11.84KK200 pKa = 9.37FMHH203 pKa = 5.21QRR205 pKa = 11.84CLKK208 pKa = 8.54GTRR211 pKa = 11.84DD212 pKa = 3.36LRR214 pKa = 11.84NIRR217 pKa = 11.84NAFNLLQGVKK227 pKa = 9.84KK228 pKa = 10.48ACQEE232 pKa = 3.62ISKK235 pKa = 10.74DD236 pKa = 4.14FILEE240 pKa = 3.98AYY242 pKa = 8.46KK243 pKa = 10.45KK244 pKa = 9.62HH245 pKa = 6.48RR246 pKa = 11.84KK247 pKa = 7.23TMEE250 pKa = 4.22TPCAGPPATEE260 pKa = 4.08LHH262 pKa = 4.77QAKK265 pKa = 10.5YY266 pKa = 10.68KK267 pKa = 9.76MLWTQGRR274 pKa = 11.84WIDD277 pKa = 2.99RR278 pKa = 11.84WKK280 pKa = 10.48KK281 pKa = 9.05YY282 pKa = 10.67KK283 pKa = 10.04PVLFKK288 pKa = 10.97PPGPAATLEE297 pKa = 4.41CPRR300 pKa = 11.84SEE302 pKa = 4.53GGGSQYY308 pKa = 11.32AFEE311 pKa = 4.2QNRR314 pKa = 11.84VFDD317 pKa = 3.8ARR319 pKa = 11.84EE320 pKa = 3.62IAVQSAIVNEE330 pKa = 4.04QLWFFFDD337 pKa = 3.41RR338 pKa = 11.84LGIRR342 pKa = 11.84DD343 pKa = 4.44AFLKK347 pKa = 7.73TQKK350 pKa = 9.65IDD352 pKa = 3.47EE353 pKa = 4.47VSPFWTWPEE362 pKa = 3.87SEE364 pKa = 4.07EE365 pKa = 4.09TGRR368 pKa = 11.84VQTVPTRR375 pKa = 11.84CLSPEE380 pKa = 3.85GKK382 pKa = 9.45IFYY385 pKa = 9.24QWQHH389 pKa = 5.73YY390 pKa = 9.18SPTWEE395 pKa = 4.18EE396 pKa = 4.33VFALYY401 pKa = 10.7NEE403 pKa = 4.44EE404 pKa = 4.14DD405 pKa = 3.69PKK407 pKa = 11.38NRR409 pKa = 11.84TVMVEE414 pKa = 4.18GIIEE418 pKa = 4.07PLKK421 pKa = 10.91LRR423 pKa = 11.84TITKK427 pKa = 9.54GPCHH431 pKa = 6.68RR432 pKa = 11.84KK433 pKa = 8.64WLSQSLQRR441 pKa = 11.84EE442 pKa = 4.17MADD445 pKa = 3.59CLDD448 pKa = 4.28GFWQFKK454 pKa = 10.0LNKK457 pKa = 9.79ANTDD461 pKa = 2.79HH462 pKa = 7.7RR463 pKa = 11.84MVTRR467 pKa = 11.84LNAACVRR474 pKa = 11.84LHH476 pKa = 6.54SKK478 pKa = 10.2QDD480 pKa = 3.05SFTPDD485 pKa = 2.9TPMWWCSGDD494 pKa = 3.69YY495 pKa = 10.59KK496 pKa = 11.0SATDD500 pKa = 4.2SISIHH505 pKa = 5.17HH506 pKa = 6.59TKK508 pKa = 10.6AALEE512 pKa = 4.07TLLNCTSPEE521 pKa = 4.05RR522 pKa = 11.84VNEE525 pKa = 3.92ACKK528 pKa = 10.25RR529 pKa = 11.84LYY531 pKa = 10.3RR532 pKa = 11.84DD533 pKa = 3.34EE534 pKa = 5.64LYY536 pKa = 10.64EE537 pKa = 5.24QIVQYY542 pKa = 9.52PKK544 pKa = 8.46WTGIPDD550 pKa = 3.51VQQVNGQLMGSVLSFPILCVINFVAYY576 pKa = 7.3WQSLEE581 pKa = 3.87EE582 pKa = 4.81HH583 pKa = 6.48YY584 pKa = 11.34GCTLNVRR591 pKa = 11.84SVPCLIHH598 pKa = 7.67GDD600 pKa = 4.08DD601 pKa = 3.9ILFKK605 pKa = 8.91TTHH608 pKa = 4.69EE609 pKa = 4.78HH610 pKa = 5.12YY611 pKa = 10.74AVWEE615 pKa = 4.07EE616 pKa = 4.12TIKK619 pKa = 11.16LFGLQKK625 pKa = 10.69SVGKK629 pKa = 10.4NYY631 pKa = 8.62FHH633 pKa = 7.28PKK635 pKa = 9.73VFTIDD640 pKa = 3.21SEE642 pKa = 4.47LWVEE646 pKa = 4.54SKK648 pKa = 10.81RR649 pKa = 11.84SDD651 pKa = 3.08GFHH654 pKa = 6.19FKK656 pKa = 10.24KK657 pKa = 10.86YY658 pKa = 10.44SGINVGSLLKK668 pKa = 10.84SKK670 pKa = 10.95VDD672 pKa = 3.49GRR674 pKa = 11.84NDD676 pKa = 3.54YY677 pKa = 10.68QRR679 pKa = 11.84LPIWDD684 pKa = 4.34KK685 pKa = 10.64FNSSIRR691 pKa = 11.84GAQDD695 pKa = 2.67KK696 pKa = 10.35EE697 pKa = 4.39LYY699 pKa = 10.12LKK701 pKa = 10.78RR702 pKa = 11.84FLHH705 pKa = 4.97YY706 pKa = 9.96HH707 pKa = 6.47KK708 pKa = 10.49STIKK712 pKa = 10.56PMTWTKK718 pKa = 11.15AGVLNLFLPRR728 pKa = 11.84EE729 pKa = 3.86RR730 pKa = 11.84GGIGFEE736 pKa = 4.82LPWKK740 pKa = 9.79PNEE743 pKa = 4.11IPIKK747 pKa = 10.54EE748 pKa = 4.27NGTPLVAVTKK758 pKa = 10.13HH759 pKa = 4.84QLNLASALCNSFRR772 pKa = 11.84EE773 pKa = 4.29AGPLKK778 pKa = 10.5SLAIVAVVDD787 pKa = 3.52KK788 pKa = 11.07TDD790 pKa = 2.91ATTEE794 pKa = 3.54KK795 pKa = 10.35RR796 pKa = 11.84YY797 pKa = 9.88RR798 pKa = 11.84LPFQEE803 pKa = 4.23VWKK806 pKa = 10.79HH807 pKa = 4.22EE808 pKa = 4.07STYY811 pKa = 11.09ARR813 pKa = 11.84EE814 pKa = 4.35IPGEE818 pKa = 3.96LADD821 pKa = 5.36PILSEE826 pKa = 4.37VPSGKK831 pKa = 9.47PEE833 pKa = 3.85EE834 pKa = 4.13YY835 pKa = 10.09RR836 pKa = 11.84LRR838 pKa = 11.84KK839 pKa = 9.62PEE841 pKa = 3.68IRR843 pKa = 11.84SLARR847 pKa = 11.84GPKK850 pKa = 8.81RR851 pKa = 11.84FRR853 pKa = 11.84VCTHH857 pKa = 6.73PPIAPNPRR865 pKa = 11.84IGRR868 pKa = 11.84IQRR871 pKa = 11.84PGEE874 pKa = 3.82LVVEE878 pKa = 4.67DD879 pKa = 4.58EE880 pKa = 4.93CVHH883 pKa = 6.58TYY885 pKa = 7.75PTEE888 pKa = 4.07IFSFPYY894 pKa = 9.75VRR896 pKa = 11.84SRR898 pKa = 11.84TIDD901 pKa = 3.32PDD903 pKa = 3.98VFWEE907 pKa = 4.58SIAWAANQYY916 pKa = 11.11NSGCSSTHH924 pKa = 5.18
Molecular weight: 106.08 kDa
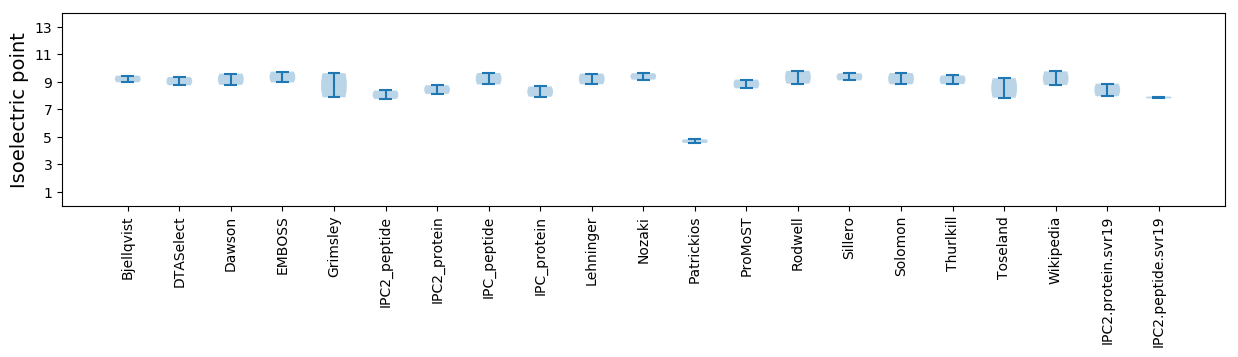
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJ26|A0A1L3KJ26_9VIRU Uncharacterized protein OS=Wenzhou narna-like virus 4 OX=1923579 PE=4 SV=1
MM1 pKa = 6.92TKK3 pKa = 10.07SRR5 pKa = 11.84RR6 pKa = 11.84GNTKK10 pKa = 9.54SGSRR14 pKa = 11.84PGTKK18 pKa = 8.95TKK20 pKa = 10.86AGLRR24 pKa = 11.84PAIAAKK30 pKa = 9.74IAKK33 pKa = 8.43PVKK36 pKa = 9.83AGRR39 pKa = 11.84FRR41 pKa = 11.84ALLNPGHH48 pKa = 6.45SPMEE52 pKa = 4.09HH53 pKa = 6.69ACAKK57 pKa = 10.32HH58 pKa = 4.14YY59 pKa = 7.68TQTVLDD65 pKa = 4.31PFNTEE70 pKa = 4.14AGACVPMLPCLDD82 pKa = 3.46SAKK85 pKa = 10.19RR86 pKa = 11.84KK87 pKa = 9.56VFARR91 pKa = 11.84GTGMTQSSNGFGGIVASTSVTKK113 pKa = 10.41DD114 pKa = 3.27APAVWYY120 pKa = 9.14TDD122 pKa = 3.25GSGTGDD128 pKa = 3.4TLTATHH134 pKa = 6.27NVAAYY139 pKa = 10.58SNSEE143 pKa = 4.19LVNTDD148 pKa = 3.07LTGGNVQARR157 pKa = 11.84IVGCGIRR164 pKa = 11.84LRR166 pKa = 11.84YY167 pKa = 9.02IGKK170 pKa = 9.69QIDD173 pKa = 3.32MSGRR177 pKa = 11.84TFALEE182 pKa = 4.18EE183 pKa = 4.58PNHH186 pKa = 6.52LNTINYY192 pKa = 7.52TVANCQALDD201 pKa = 3.38RR202 pKa = 11.84CKK204 pKa = 10.77SKK206 pKa = 10.84NLSRR210 pKa = 11.84EE211 pKa = 3.79WTVACWQPVRR221 pKa = 11.84PGEE224 pKa = 4.36QSYY227 pKa = 10.71QMNANAATDD236 pKa = 3.82PVHH239 pKa = 5.78YY240 pKa = 10.45QPLCVLIQAQAGQSLPFEE258 pKa = 4.76WEE260 pKa = 4.01WFCHH264 pKa = 5.75YY265 pKa = 10.44EE266 pKa = 4.3AIGYY270 pKa = 7.43GARR273 pKa = 11.84GKK275 pKa = 10.27SRR277 pKa = 11.84SHH279 pKa = 5.83VAPIAGPKK287 pKa = 9.96AIAALQQTPSDD298 pKa = 3.91VFDD301 pKa = 4.45DD302 pKa = 4.01VSNKK306 pKa = 9.75RR307 pKa = 11.84LSSGRR312 pKa = 11.84LADD315 pKa = 3.74NMISQGSSFDD325 pKa = 3.32WSKK328 pKa = 11.1LADD331 pKa = 3.32TGLRR335 pKa = 11.84VAQTAIGAVTSRR347 pKa = 11.84AAAQYY352 pKa = 9.9MSGGLAALALL362 pKa = 4.24
MM1 pKa = 6.92TKK3 pKa = 10.07SRR5 pKa = 11.84RR6 pKa = 11.84GNTKK10 pKa = 9.54SGSRR14 pKa = 11.84PGTKK18 pKa = 8.95TKK20 pKa = 10.86AGLRR24 pKa = 11.84PAIAAKK30 pKa = 9.74IAKK33 pKa = 8.43PVKK36 pKa = 9.83AGRR39 pKa = 11.84FRR41 pKa = 11.84ALLNPGHH48 pKa = 6.45SPMEE52 pKa = 4.09HH53 pKa = 6.69ACAKK57 pKa = 10.32HH58 pKa = 4.14YY59 pKa = 7.68TQTVLDD65 pKa = 4.31PFNTEE70 pKa = 4.14AGACVPMLPCLDD82 pKa = 3.46SAKK85 pKa = 10.19RR86 pKa = 11.84KK87 pKa = 9.56VFARR91 pKa = 11.84GTGMTQSSNGFGGIVASTSVTKK113 pKa = 10.41DD114 pKa = 3.27APAVWYY120 pKa = 9.14TDD122 pKa = 3.25GSGTGDD128 pKa = 3.4TLTATHH134 pKa = 6.27NVAAYY139 pKa = 10.58SNSEE143 pKa = 4.19LVNTDD148 pKa = 3.07LTGGNVQARR157 pKa = 11.84IVGCGIRR164 pKa = 11.84LRR166 pKa = 11.84YY167 pKa = 9.02IGKK170 pKa = 9.69QIDD173 pKa = 3.32MSGRR177 pKa = 11.84TFALEE182 pKa = 4.18EE183 pKa = 4.58PNHH186 pKa = 6.52LNTINYY192 pKa = 7.52TVANCQALDD201 pKa = 3.38RR202 pKa = 11.84CKK204 pKa = 10.77SKK206 pKa = 10.84NLSRR210 pKa = 11.84EE211 pKa = 3.79WTVACWQPVRR221 pKa = 11.84PGEE224 pKa = 4.36QSYY227 pKa = 10.71QMNANAATDD236 pKa = 3.82PVHH239 pKa = 5.78YY240 pKa = 10.45QPLCVLIQAQAGQSLPFEE258 pKa = 4.76WEE260 pKa = 4.01WFCHH264 pKa = 5.75YY265 pKa = 10.44EE266 pKa = 4.3AIGYY270 pKa = 7.43GARR273 pKa = 11.84GKK275 pKa = 10.27SRR277 pKa = 11.84SHH279 pKa = 5.83VAPIAGPKK287 pKa = 9.96AIAALQQTPSDD298 pKa = 3.91VFDD301 pKa = 4.45DD302 pKa = 4.01VSNKK306 pKa = 9.75RR307 pKa = 11.84LSSGRR312 pKa = 11.84LADD315 pKa = 3.74NMISQGSSFDD325 pKa = 3.32WSKK328 pKa = 11.1LADD331 pKa = 3.32TGLRR335 pKa = 11.84VAQTAIGAVTSRR347 pKa = 11.84AAAQYY352 pKa = 9.9MSGGLAALALL362 pKa = 4.24
Molecular weight: 38.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1286 |
362 |
924 |
643.0 |
72.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.932 ± 2.602 | 2.411 ± 0.039 |
4.277 ± 0.069 | 5.754 ± 1.541 |
4.121 ± 0.842 | 6.454 ± 1.371 |
2.488 ± 0.143 | 4.821 ± 0.491 |
6.687 ± 0.883 | 7.854 ± 0.488 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.555 ± 0.337 | 3.81 ± 0.456 |
6.143 ± 0.603 | 4.432 ± 0.136 |
5.988 ± 0.096 | 7.154 ± 0.441 |
6.765 ± 0.357 | 5.677 ± 0.064 |
2.566 ± 0.468 | 3.11 ± 0.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
