
Rhizophagus irregularis mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 9.28
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385HDR8|A0A385HDR8_9VIRU RNA-dependent RNA polymerase OS=Rhizophagus irregularis mitovirus 1 OX=2320186 PE=4 SV=1
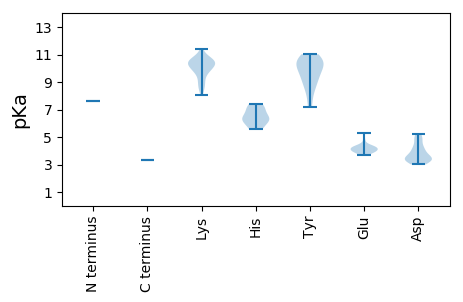
MM1 pKa = 7.62KK2 pKa = 10.56NLFQNFVEE10 pKa = 4.44RR11 pKa = 11.84KK12 pKa = 8.48DD13 pKa = 3.3RR14 pKa = 11.84ARR16 pKa = 11.84IEE18 pKa = 4.01KK19 pKa = 10.51LGTGVGNYY27 pKa = 9.04VSLIIRR33 pKa = 11.84FGYY36 pKa = 10.67AIAFLLGGRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 5.63ARR49 pKa = 11.84FYY51 pKa = 11.02LAVVHH56 pKa = 5.56YY57 pKa = 8.66GKK59 pKa = 9.43WLRR62 pKa = 11.84EE63 pKa = 3.9WNGSNKK69 pKa = 10.25ALVKK73 pKa = 10.15YY74 pKa = 10.48LKK76 pKa = 10.11ACSVTLMQFAGQGPIASSRR95 pKa = 11.84DD96 pKa = 3.22LGGVHH101 pKa = 6.27GRR103 pKa = 11.84SRR105 pKa = 11.84SNLPRR110 pKa = 11.84VIPVYY115 pKa = 9.97HH116 pKa = 6.31RR117 pKa = 11.84VGIRR121 pKa = 11.84RR122 pKa = 11.84GDD124 pKa = 3.33SRR126 pKa = 11.84IIQFWLSLLGVYY138 pKa = 10.17RR139 pKa = 11.84IIEE142 pKa = 4.18CKK144 pKa = 10.57GRR146 pKa = 11.84VSYY149 pKa = 11.01KK150 pKa = 10.6SITQPGVEE158 pKa = 3.95RR159 pKa = 11.84QFKK162 pKa = 9.48VVYY165 pKa = 9.07EE166 pKa = 4.28LGVMLQSLKK175 pKa = 10.27TSDD178 pKa = 3.0RR179 pKa = 11.84TAQRR183 pKa = 11.84VLDD186 pKa = 4.93PIDD189 pKa = 4.33LKK191 pKa = 11.38SEE193 pKa = 4.33LKK195 pKa = 9.71FQPRR199 pKa = 11.84ALLTSGANVPGGVGGFWALAWDD221 pKa = 3.98ALKK224 pKa = 10.03IWKK227 pKa = 9.14VRR229 pKa = 11.84EE230 pKa = 4.16TPFGGAVNAFAYY242 pKa = 8.24LTGQLDD248 pKa = 4.15LLGLIEE254 pKa = 4.8LAATTATHH262 pKa = 7.41DD263 pKa = 3.55FRR265 pKa = 11.84NPEE268 pKa = 4.0SVQDD272 pKa = 3.65KK273 pKa = 10.65RR274 pKa = 11.84VGSKK278 pKa = 10.69DD279 pKa = 3.25SVMGVQKK286 pKa = 10.9GFATLFGKK294 pKa = 8.98MNKK297 pKa = 8.8TGEE300 pKa = 4.32DD301 pKa = 3.43RR302 pKa = 11.84SCTQWRR308 pKa = 11.84MAPLSNFVTNLVLGRR323 pKa = 11.84LHH325 pKa = 7.07VIPEE329 pKa = 4.13PAGKK333 pKa = 8.62MRR335 pKa = 11.84VVAMGTWWVQCMLYY349 pKa = 9.69PLHH352 pKa = 7.2RR353 pKa = 11.84ILYY356 pKa = 8.54RR357 pKa = 11.84KK358 pKa = 9.95LGLIPNDD365 pKa = 4.02GTWDD369 pKa = 3.34QSKK372 pKa = 9.71PLEE375 pKa = 4.29GMAAKK380 pKa = 10.23VKK382 pKa = 10.39EE383 pKa = 4.14ILSSGGIPQVFSYY396 pKa = 10.77DD397 pKa = 3.23LSAATDD403 pKa = 4.28RR404 pKa = 11.84FPVWYY409 pKa = 9.3QVEE412 pKa = 4.31VLAFLTNRR420 pKa = 11.84RR421 pKa = 11.84FAEE424 pKa = 4.09TWRR427 pKa = 11.84DD428 pKa = 3.5LLIMPRR434 pKa = 11.84YY435 pKa = 7.18YY436 pKa = 10.18TGSITVIPRR445 pKa = 11.84GDD447 pKa = 3.39ALVYY451 pKa = 10.8GSGQPMGLYY460 pKa = 10.35SSWAMFSLAHH470 pKa = 6.45HH471 pKa = 6.81LLVQQAASRR480 pKa = 11.84VGYY483 pKa = 9.71KK484 pKa = 9.87GWYY487 pKa = 7.39PWYY490 pKa = 10.41ALLGDD495 pKa = 5.2DD496 pKa = 4.96IVILGEE502 pKa = 4.27DD503 pKa = 3.23VAGAYY508 pKa = 9.36KK509 pKa = 10.29DD510 pKa = 4.79LCDD513 pKa = 3.65QLQVKK518 pKa = 10.12IGLAKK523 pKa = 10.55SLISSNGSFEE533 pKa = 3.92FAKK536 pKa = 10.16RR537 pKa = 11.84YY538 pKa = 8.86YY539 pKa = 10.87YY540 pKa = 10.45KK541 pKa = 11.12GNDD544 pKa = 3.63CSPVSIRR551 pKa = 11.84EE552 pKa = 3.93YY553 pKa = 9.68WVSLSSLPAFAEE565 pKa = 3.69MVMRR569 pKa = 11.84IKK571 pKa = 10.66RR572 pKa = 11.84SVPDD576 pKa = 3.26IRR578 pKa = 11.84LSDD581 pKa = 3.4AVRR584 pKa = 11.84AYY586 pKa = 9.86KK587 pKa = 10.4YY588 pKa = 10.27GYY590 pKa = 10.12HH591 pKa = 6.19SVAKK595 pKa = 9.05LTQCIVKK602 pKa = 10.38LGNSRR607 pKa = 11.84LANLLAILMLPGGPFEE623 pKa = 5.31RR624 pKa = 11.84SLLSLFSPTSTAVRR638 pKa = 11.84PNDD641 pKa = 3.46NVVDD645 pKa = 4.06SPITEE650 pKa = 3.76RR651 pKa = 11.84RR652 pKa = 11.84VKK654 pKa = 10.49SVSRR658 pKa = 11.84TLGQSLCSVSARR670 pKa = 11.84ASSYY674 pKa = 10.8LSSLTVYY681 pKa = 10.36EE682 pKa = 4.0NEE684 pKa = 4.23LRR686 pKa = 11.84VFDD689 pKa = 5.09PIGFLRR695 pKa = 11.84SVVISRR701 pKa = 11.84MRR703 pKa = 11.84VTLTNSSHH711 pKa = 6.22VGLLEE716 pKa = 3.88RR717 pKa = 11.84FGRR720 pKa = 11.84YY721 pKa = 9.36LLDD724 pKa = 4.02GKK726 pKa = 10.84LNSRR730 pKa = 11.84GLVTLLGRR738 pKa = 11.84IIPIWKK744 pKa = 9.91FSVGDD749 pKa = 3.47VAAYY753 pKa = 8.05PDD755 pKa = 4.6PFALDD760 pKa = 3.31AAGGVLARR768 pKa = 11.84PQTTKK773 pKa = 10.52FLKK776 pKa = 10.55LRR778 pKa = 11.84VRR780 pKa = 11.84LLGLPWKK787 pKa = 9.8GRR789 pKa = 11.84PKK791 pKa = 10.58GGPKK795 pKa = 8.55VTRR798 pKa = 11.84KK799 pKa = 9.7GSKK802 pKa = 10.1ARR804 pKa = 11.84MGRR807 pKa = 11.84KK808 pKa = 8.09LTTT811 pKa = 3.35
MM1 pKa = 7.62KK2 pKa = 10.56NLFQNFVEE10 pKa = 4.44RR11 pKa = 11.84KK12 pKa = 8.48DD13 pKa = 3.3RR14 pKa = 11.84ARR16 pKa = 11.84IEE18 pKa = 4.01KK19 pKa = 10.51LGTGVGNYY27 pKa = 9.04VSLIIRR33 pKa = 11.84FGYY36 pKa = 10.67AIAFLLGGRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 5.63ARR49 pKa = 11.84FYY51 pKa = 11.02LAVVHH56 pKa = 5.56YY57 pKa = 8.66GKK59 pKa = 9.43WLRR62 pKa = 11.84EE63 pKa = 3.9WNGSNKK69 pKa = 10.25ALVKK73 pKa = 10.15YY74 pKa = 10.48LKK76 pKa = 10.11ACSVTLMQFAGQGPIASSRR95 pKa = 11.84DD96 pKa = 3.22LGGVHH101 pKa = 6.27GRR103 pKa = 11.84SRR105 pKa = 11.84SNLPRR110 pKa = 11.84VIPVYY115 pKa = 9.97HH116 pKa = 6.31RR117 pKa = 11.84VGIRR121 pKa = 11.84RR122 pKa = 11.84GDD124 pKa = 3.33SRR126 pKa = 11.84IIQFWLSLLGVYY138 pKa = 10.17RR139 pKa = 11.84IIEE142 pKa = 4.18CKK144 pKa = 10.57GRR146 pKa = 11.84VSYY149 pKa = 11.01KK150 pKa = 10.6SITQPGVEE158 pKa = 3.95RR159 pKa = 11.84QFKK162 pKa = 9.48VVYY165 pKa = 9.07EE166 pKa = 4.28LGVMLQSLKK175 pKa = 10.27TSDD178 pKa = 3.0RR179 pKa = 11.84TAQRR183 pKa = 11.84VLDD186 pKa = 4.93PIDD189 pKa = 4.33LKK191 pKa = 11.38SEE193 pKa = 4.33LKK195 pKa = 9.71FQPRR199 pKa = 11.84ALLTSGANVPGGVGGFWALAWDD221 pKa = 3.98ALKK224 pKa = 10.03IWKK227 pKa = 9.14VRR229 pKa = 11.84EE230 pKa = 4.16TPFGGAVNAFAYY242 pKa = 8.24LTGQLDD248 pKa = 4.15LLGLIEE254 pKa = 4.8LAATTATHH262 pKa = 7.41DD263 pKa = 3.55FRR265 pKa = 11.84NPEE268 pKa = 4.0SVQDD272 pKa = 3.65KK273 pKa = 10.65RR274 pKa = 11.84VGSKK278 pKa = 10.69DD279 pKa = 3.25SVMGVQKK286 pKa = 10.9GFATLFGKK294 pKa = 8.98MNKK297 pKa = 8.8TGEE300 pKa = 4.32DD301 pKa = 3.43RR302 pKa = 11.84SCTQWRR308 pKa = 11.84MAPLSNFVTNLVLGRR323 pKa = 11.84LHH325 pKa = 7.07VIPEE329 pKa = 4.13PAGKK333 pKa = 8.62MRR335 pKa = 11.84VVAMGTWWVQCMLYY349 pKa = 9.69PLHH352 pKa = 7.2RR353 pKa = 11.84ILYY356 pKa = 8.54RR357 pKa = 11.84KK358 pKa = 9.95LGLIPNDD365 pKa = 4.02GTWDD369 pKa = 3.34QSKK372 pKa = 9.71PLEE375 pKa = 4.29GMAAKK380 pKa = 10.23VKK382 pKa = 10.39EE383 pKa = 4.14ILSSGGIPQVFSYY396 pKa = 10.77DD397 pKa = 3.23LSAATDD403 pKa = 4.28RR404 pKa = 11.84FPVWYY409 pKa = 9.3QVEE412 pKa = 4.31VLAFLTNRR420 pKa = 11.84RR421 pKa = 11.84FAEE424 pKa = 4.09TWRR427 pKa = 11.84DD428 pKa = 3.5LLIMPRR434 pKa = 11.84YY435 pKa = 7.18YY436 pKa = 10.18TGSITVIPRR445 pKa = 11.84GDD447 pKa = 3.39ALVYY451 pKa = 10.8GSGQPMGLYY460 pKa = 10.35SSWAMFSLAHH470 pKa = 6.45HH471 pKa = 6.81LLVQQAASRR480 pKa = 11.84VGYY483 pKa = 9.71KK484 pKa = 9.87GWYY487 pKa = 7.39PWYY490 pKa = 10.41ALLGDD495 pKa = 5.2DD496 pKa = 4.96IVILGEE502 pKa = 4.27DD503 pKa = 3.23VAGAYY508 pKa = 9.36KK509 pKa = 10.29DD510 pKa = 4.79LCDD513 pKa = 3.65QLQVKK518 pKa = 10.12IGLAKK523 pKa = 10.55SLISSNGSFEE533 pKa = 3.92FAKK536 pKa = 10.16RR537 pKa = 11.84YY538 pKa = 8.86YY539 pKa = 10.87YY540 pKa = 10.45KK541 pKa = 11.12GNDD544 pKa = 3.63CSPVSIRR551 pKa = 11.84EE552 pKa = 3.93YY553 pKa = 9.68WVSLSSLPAFAEE565 pKa = 3.69MVMRR569 pKa = 11.84IKK571 pKa = 10.66RR572 pKa = 11.84SVPDD576 pKa = 3.26IRR578 pKa = 11.84LSDD581 pKa = 3.4AVRR584 pKa = 11.84AYY586 pKa = 9.86KK587 pKa = 10.4YY588 pKa = 10.27GYY590 pKa = 10.12HH591 pKa = 6.19SVAKK595 pKa = 9.05LTQCIVKK602 pKa = 10.38LGNSRR607 pKa = 11.84LANLLAILMLPGGPFEE623 pKa = 5.31RR624 pKa = 11.84SLLSLFSPTSTAVRR638 pKa = 11.84PNDD641 pKa = 3.46NVVDD645 pKa = 4.06SPITEE650 pKa = 3.76RR651 pKa = 11.84RR652 pKa = 11.84VKK654 pKa = 10.49SVSRR658 pKa = 11.84TLGQSLCSVSARR670 pKa = 11.84ASSYY674 pKa = 10.8LSSLTVYY681 pKa = 10.36EE682 pKa = 4.0NEE684 pKa = 4.23LRR686 pKa = 11.84VFDD689 pKa = 5.09PIGFLRR695 pKa = 11.84SVVISRR701 pKa = 11.84MRR703 pKa = 11.84VTLTNSSHH711 pKa = 6.22VGLLEE716 pKa = 3.88RR717 pKa = 11.84FGRR720 pKa = 11.84YY721 pKa = 9.36LLDD724 pKa = 4.02GKK726 pKa = 10.84LNSRR730 pKa = 11.84GLVTLLGRR738 pKa = 11.84IIPIWKK744 pKa = 9.91FSVGDD749 pKa = 3.47VAAYY753 pKa = 8.05PDD755 pKa = 4.6PFALDD760 pKa = 3.31AAGGVLARR768 pKa = 11.84PQTTKK773 pKa = 10.52FLKK776 pKa = 10.55LRR778 pKa = 11.84VRR780 pKa = 11.84LLGLPWKK787 pKa = 9.8GRR789 pKa = 11.84PKK791 pKa = 10.58GGPKK795 pKa = 8.55VTRR798 pKa = 11.84KK799 pKa = 9.7GSKK802 pKa = 10.1ARR804 pKa = 11.84MGRR807 pKa = 11.84KK808 pKa = 8.09LTTT811 pKa = 3.35
Molecular weight: 90.46 kDa
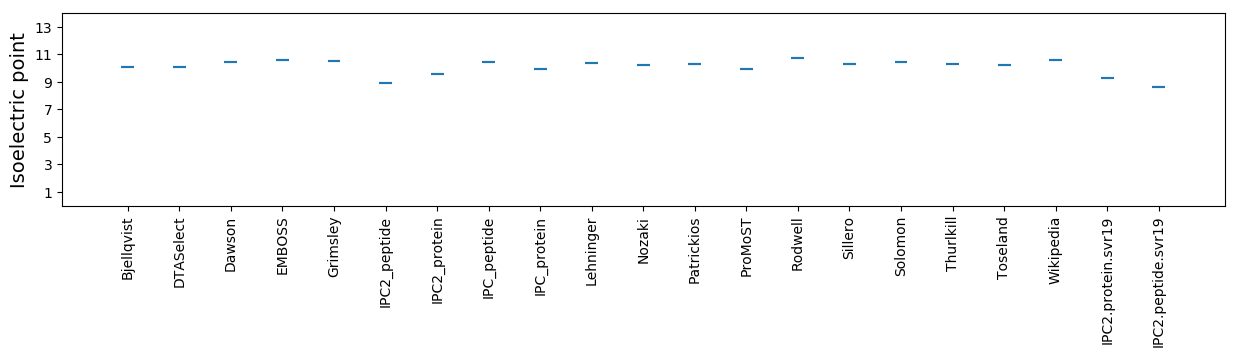
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385HDR8|A0A385HDR8_9VIRU RNA-dependent RNA polymerase OS=Rhizophagus irregularis mitovirus 1 OX=2320186 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.56NLFQNFVEE10 pKa = 4.44RR11 pKa = 11.84KK12 pKa = 8.48DD13 pKa = 3.3RR14 pKa = 11.84ARR16 pKa = 11.84IEE18 pKa = 4.01KK19 pKa = 10.51LGTGVGNYY27 pKa = 9.04VSLIIRR33 pKa = 11.84FGYY36 pKa = 10.67AIAFLLGGRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 5.63ARR49 pKa = 11.84FYY51 pKa = 11.02LAVVHH56 pKa = 5.56YY57 pKa = 8.66GKK59 pKa = 9.43WLRR62 pKa = 11.84EE63 pKa = 3.9WNGSNKK69 pKa = 10.25ALVKK73 pKa = 10.15YY74 pKa = 10.48LKK76 pKa = 10.11ACSVTLMQFAGQGPIASSRR95 pKa = 11.84DD96 pKa = 3.22LGGVHH101 pKa = 6.27GRR103 pKa = 11.84SRR105 pKa = 11.84SNLPRR110 pKa = 11.84VIPVYY115 pKa = 9.97HH116 pKa = 6.31RR117 pKa = 11.84VGIRR121 pKa = 11.84RR122 pKa = 11.84GDD124 pKa = 3.33SRR126 pKa = 11.84IIQFWLSLLGVYY138 pKa = 10.17RR139 pKa = 11.84IIEE142 pKa = 4.18CKK144 pKa = 10.57GRR146 pKa = 11.84VSYY149 pKa = 11.01KK150 pKa = 10.6SITQPGVEE158 pKa = 3.95RR159 pKa = 11.84QFKK162 pKa = 9.48VVYY165 pKa = 9.07EE166 pKa = 4.28LGVMLQSLKK175 pKa = 10.27TSDD178 pKa = 3.0RR179 pKa = 11.84TAQRR183 pKa = 11.84VLDD186 pKa = 4.93PIDD189 pKa = 4.33LKK191 pKa = 11.38SEE193 pKa = 4.33LKK195 pKa = 9.71FQPRR199 pKa = 11.84ALLTSGANVPGGVGGFWALAWDD221 pKa = 3.98ALKK224 pKa = 10.03IWKK227 pKa = 9.14VRR229 pKa = 11.84EE230 pKa = 4.16TPFGGAVNAFAYY242 pKa = 8.24LTGQLDD248 pKa = 4.15LLGLIEE254 pKa = 4.8LAATTATHH262 pKa = 7.41DD263 pKa = 3.55FRR265 pKa = 11.84NPEE268 pKa = 4.0SVQDD272 pKa = 3.65KK273 pKa = 10.65RR274 pKa = 11.84VGSKK278 pKa = 10.69DD279 pKa = 3.25SVMGVQKK286 pKa = 10.9GFATLFGKK294 pKa = 8.98MNKK297 pKa = 8.8TGEE300 pKa = 4.32DD301 pKa = 3.43RR302 pKa = 11.84SCTQWRR308 pKa = 11.84MAPLSNFVTNLVLGRR323 pKa = 11.84LHH325 pKa = 7.07VIPEE329 pKa = 4.13PAGKK333 pKa = 8.62MRR335 pKa = 11.84VVAMGTWWVQCMLYY349 pKa = 9.69PLHH352 pKa = 7.2RR353 pKa = 11.84ILYY356 pKa = 8.54RR357 pKa = 11.84KK358 pKa = 9.95LGLIPNDD365 pKa = 4.02GTWDD369 pKa = 3.34QSKK372 pKa = 9.71PLEE375 pKa = 4.29GMAAKK380 pKa = 10.23VKK382 pKa = 10.39EE383 pKa = 4.14ILSSGGIPQVFSYY396 pKa = 10.77DD397 pKa = 3.23LSAATDD403 pKa = 4.28RR404 pKa = 11.84FPVWYY409 pKa = 9.3QVEE412 pKa = 4.31VLAFLTNRR420 pKa = 11.84RR421 pKa = 11.84FAEE424 pKa = 4.09TWRR427 pKa = 11.84DD428 pKa = 3.5LLIMPRR434 pKa = 11.84YY435 pKa = 7.18YY436 pKa = 10.18TGSITVIPRR445 pKa = 11.84GDD447 pKa = 3.39ALVYY451 pKa = 10.8GSGQPMGLYY460 pKa = 10.35SSWAMFSLAHH470 pKa = 6.45HH471 pKa = 6.81LLVQQAASRR480 pKa = 11.84VGYY483 pKa = 9.71KK484 pKa = 9.87GWYY487 pKa = 7.39PWYY490 pKa = 10.41ALLGDD495 pKa = 5.2DD496 pKa = 4.96IVILGEE502 pKa = 4.27DD503 pKa = 3.23VAGAYY508 pKa = 9.36KK509 pKa = 10.29DD510 pKa = 4.79LCDD513 pKa = 3.65QLQVKK518 pKa = 10.12IGLAKK523 pKa = 10.55SLISSNGSFEE533 pKa = 3.92FAKK536 pKa = 10.16RR537 pKa = 11.84YY538 pKa = 8.86YY539 pKa = 10.87YY540 pKa = 10.45KK541 pKa = 11.12GNDD544 pKa = 3.63CSPVSIRR551 pKa = 11.84EE552 pKa = 3.93YY553 pKa = 9.68WVSLSSLPAFAEE565 pKa = 3.69MVMRR569 pKa = 11.84IKK571 pKa = 10.66RR572 pKa = 11.84SVPDD576 pKa = 3.26IRR578 pKa = 11.84LSDD581 pKa = 3.4AVRR584 pKa = 11.84AYY586 pKa = 9.86KK587 pKa = 10.4YY588 pKa = 10.27GYY590 pKa = 10.12HH591 pKa = 6.19SVAKK595 pKa = 9.05LTQCIVKK602 pKa = 10.38LGNSRR607 pKa = 11.84LANLLAILMLPGGPFEE623 pKa = 5.31RR624 pKa = 11.84SLLSLFSPTSTAVRR638 pKa = 11.84PNDD641 pKa = 3.46NVVDD645 pKa = 4.06SPITEE650 pKa = 3.76RR651 pKa = 11.84RR652 pKa = 11.84VKK654 pKa = 10.49SVSRR658 pKa = 11.84TLGQSLCSVSARR670 pKa = 11.84ASSYY674 pKa = 10.8LSSLTVYY681 pKa = 10.36EE682 pKa = 4.0NEE684 pKa = 4.23LRR686 pKa = 11.84VFDD689 pKa = 5.09PIGFLRR695 pKa = 11.84SVVISRR701 pKa = 11.84MRR703 pKa = 11.84VTLTNSSHH711 pKa = 6.22VGLLEE716 pKa = 3.88RR717 pKa = 11.84FGRR720 pKa = 11.84YY721 pKa = 9.36LLDD724 pKa = 4.02GKK726 pKa = 10.84LNSRR730 pKa = 11.84GLVTLLGRR738 pKa = 11.84IIPIWKK744 pKa = 9.91FSVGDD749 pKa = 3.47VAAYY753 pKa = 8.05PDD755 pKa = 4.6PFALDD760 pKa = 3.31AAGGVLARR768 pKa = 11.84PQTTKK773 pKa = 10.52FLKK776 pKa = 10.55LRR778 pKa = 11.84VRR780 pKa = 11.84LLGLPWKK787 pKa = 9.8GRR789 pKa = 11.84PKK791 pKa = 10.58GGPKK795 pKa = 8.55VTRR798 pKa = 11.84KK799 pKa = 9.7GSKK802 pKa = 10.1ARR804 pKa = 11.84MGRR807 pKa = 11.84KK808 pKa = 8.09LTTT811 pKa = 3.35
MM1 pKa = 7.62KK2 pKa = 10.56NLFQNFVEE10 pKa = 4.44RR11 pKa = 11.84KK12 pKa = 8.48DD13 pKa = 3.3RR14 pKa = 11.84ARR16 pKa = 11.84IEE18 pKa = 4.01KK19 pKa = 10.51LGTGVGNYY27 pKa = 9.04VSLIIRR33 pKa = 11.84FGYY36 pKa = 10.67AIAFLLGGRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 5.63ARR49 pKa = 11.84FYY51 pKa = 11.02LAVVHH56 pKa = 5.56YY57 pKa = 8.66GKK59 pKa = 9.43WLRR62 pKa = 11.84EE63 pKa = 3.9WNGSNKK69 pKa = 10.25ALVKK73 pKa = 10.15YY74 pKa = 10.48LKK76 pKa = 10.11ACSVTLMQFAGQGPIASSRR95 pKa = 11.84DD96 pKa = 3.22LGGVHH101 pKa = 6.27GRR103 pKa = 11.84SRR105 pKa = 11.84SNLPRR110 pKa = 11.84VIPVYY115 pKa = 9.97HH116 pKa = 6.31RR117 pKa = 11.84VGIRR121 pKa = 11.84RR122 pKa = 11.84GDD124 pKa = 3.33SRR126 pKa = 11.84IIQFWLSLLGVYY138 pKa = 10.17RR139 pKa = 11.84IIEE142 pKa = 4.18CKK144 pKa = 10.57GRR146 pKa = 11.84VSYY149 pKa = 11.01KK150 pKa = 10.6SITQPGVEE158 pKa = 3.95RR159 pKa = 11.84QFKK162 pKa = 9.48VVYY165 pKa = 9.07EE166 pKa = 4.28LGVMLQSLKK175 pKa = 10.27TSDD178 pKa = 3.0RR179 pKa = 11.84TAQRR183 pKa = 11.84VLDD186 pKa = 4.93PIDD189 pKa = 4.33LKK191 pKa = 11.38SEE193 pKa = 4.33LKK195 pKa = 9.71FQPRR199 pKa = 11.84ALLTSGANVPGGVGGFWALAWDD221 pKa = 3.98ALKK224 pKa = 10.03IWKK227 pKa = 9.14VRR229 pKa = 11.84EE230 pKa = 4.16TPFGGAVNAFAYY242 pKa = 8.24LTGQLDD248 pKa = 4.15LLGLIEE254 pKa = 4.8LAATTATHH262 pKa = 7.41DD263 pKa = 3.55FRR265 pKa = 11.84NPEE268 pKa = 4.0SVQDD272 pKa = 3.65KK273 pKa = 10.65RR274 pKa = 11.84VGSKK278 pKa = 10.69DD279 pKa = 3.25SVMGVQKK286 pKa = 10.9GFATLFGKK294 pKa = 8.98MNKK297 pKa = 8.8TGEE300 pKa = 4.32DD301 pKa = 3.43RR302 pKa = 11.84SCTQWRR308 pKa = 11.84MAPLSNFVTNLVLGRR323 pKa = 11.84LHH325 pKa = 7.07VIPEE329 pKa = 4.13PAGKK333 pKa = 8.62MRR335 pKa = 11.84VVAMGTWWVQCMLYY349 pKa = 9.69PLHH352 pKa = 7.2RR353 pKa = 11.84ILYY356 pKa = 8.54RR357 pKa = 11.84KK358 pKa = 9.95LGLIPNDD365 pKa = 4.02GTWDD369 pKa = 3.34QSKK372 pKa = 9.71PLEE375 pKa = 4.29GMAAKK380 pKa = 10.23VKK382 pKa = 10.39EE383 pKa = 4.14ILSSGGIPQVFSYY396 pKa = 10.77DD397 pKa = 3.23LSAATDD403 pKa = 4.28RR404 pKa = 11.84FPVWYY409 pKa = 9.3QVEE412 pKa = 4.31VLAFLTNRR420 pKa = 11.84RR421 pKa = 11.84FAEE424 pKa = 4.09TWRR427 pKa = 11.84DD428 pKa = 3.5LLIMPRR434 pKa = 11.84YY435 pKa = 7.18YY436 pKa = 10.18TGSITVIPRR445 pKa = 11.84GDD447 pKa = 3.39ALVYY451 pKa = 10.8GSGQPMGLYY460 pKa = 10.35SSWAMFSLAHH470 pKa = 6.45HH471 pKa = 6.81LLVQQAASRR480 pKa = 11.84VGYY483 pKa = 9.71KK484 pKa = 9.87GWYY487 pKa = 7.39PWYY490 pKa = 10.41ALLGDD495 pKa = 5.2DD496 pKa = 4.96IVILGEE502 pKa = 4.27DD503 pKa = 3.23VAGAYY508 pKa = 9.36KK509 pKa = 10.29DD510 pKa = 4.79LCDD513 pKa = 3.65QLQVKK518 pKa = 10.12IGLAKK523 pKa = 10.55SLISSNGSFEE533 pKa = 3.92FAKK536 pKa = 10.16RR537 pKa = 11.84YY538 pKa = 8.86YY539 pKa = 10.87YY540 pKa = 10.45KK541 pKa = 11.12GNDD544 pKa = 3.63CSPVSIRR551 pKa = 11.84EE552 pKa = 3.93YY553 pKa = 9.68WVSLSSLPAFAEE565 pKa = 3.69MVMRR569 pKa = 11.84IKK571 pKa = 10.66RR572 pKa = 11.84SVPDD576 pKa = 3.26IRR578 pKa = 11.84LSDD581 pKa = 3.4AVRR584 pKa = 11.84AYY586 pKa = 9.86KK587 pKa = 10.4YY588 pKa = 10.27GYY590 pKa = 10.12HH591 pKa = 6.19SVAKK595 pKa = 9.05LTQCIVKK602 pKa = 10.38LGNSRR607 pKa = 11.84LANLLAILMLPGGPFEE623 pKa = 5.31RR624 pKa = 11.84SLLSLFSPTSTAVRR638 pKa = 11.84PNDD641 pKa = 3.46NVVDD645 pKa = 4.06SPITEE650 pKa = 3.76RR651 pKa = 11.84RR652 pKa = 11.84VKK654 pKa = 10.49SVSRR658 pKa = 11.84TLGQSLCSVSARR670 pKa = 11.84ASSYY674 pKa = 10.8LSSLTVYY681 pKa = 10.36EE682 pKa = 4.0NEE684 pKa = 4.23LRR686 pKa = 11.84VFDD689 pKa = 5.09PIGFLRR695 pKa = 11.84SVVISRR701 pKa = 11.84MRR703 pKa = 11.84VTLTNSSHH711 pKa = 6.22VGLLEE716 pKa = 3.88RR717 pKa = 11.84FGRR720 pKa = 11.84YY721 pKa = 9.36LLDD724 pKa = 4.02GKK726 pKa = 10.84LNSRR730 pKa = 11.84GLVTLLGRR738 pKa = 11.84IIPIWKK744 pKa = 9.91FSVGDD749 pKa = 3.47VAAYY753 pKa = 8.05PDD755 pKa = 4.6PFALDD760 pKa = 3.31AAGGVLARR768 pKa = 11.84PQTTKK773 pKa = 10.52FLKK776 pKa = 10.55LRR778 pKa = 11.84VRR780 pKa = 11.84LLGLPWKK787 pKa = 9.8GRR789 pKa = 11.84PKK791 pKa = 10.58GGPKK795 pKa = 8.55VTRR798 pKa = 11.84KK799 pKa = 9.7GSKK802 pKa = 10.1ARR804 pKa = 11.84MGRR807 pKa = 11.84KK808 pKa = 8.09LTTT811 pKa = 3.35
Molecular weight: 90.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
811 |
811 |
811 |
811.0 |
90.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.152 ± 0.0 | 0.986 ± 0.0 |
4.069 ± 0.0 | 3.083 ± 0.0 |
3.946 ± 0.0 | 9.125 ± 0.0 |
1.356 ± 0.0 | 4.686 ± 0.0 |
5.672 ± 0.0 | 11.837 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.219 ± 0.0 | 2.836 ± 0.0 |
4.562 ± 0.0 | 3.083 ± 0.0 |
8.138 ± 0.0 | 8.015 ± 0.0 |
4.439 ± 0.0 | 8.508 ± 0.0 |
2.219 ± 0.0 | 4.069 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
