
Wood mouse herpesvirus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Rhadinovirus; Murid gammaherpesvirus 7
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
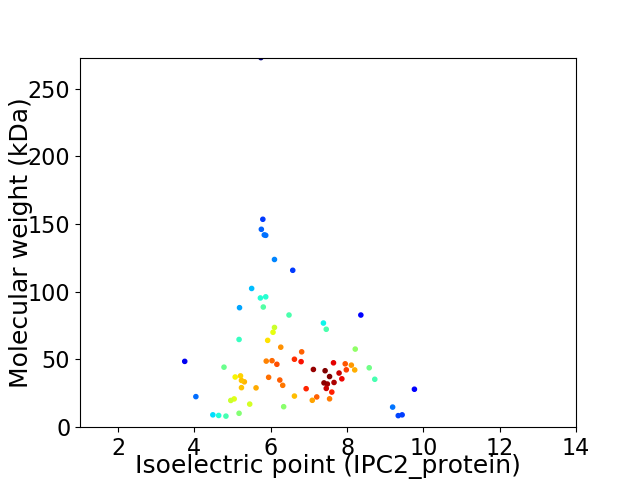
Virtual 2D-PAGE plot for 74 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0U1P1|D0U1P1_9GAMA Virion protein G52 OS=Wood mouse herpesvirus OX=432370 GN=ORF52 PE=1 SV=1
MM1 pKa = 7.57CGVKK5 pKa = 10.28PIVKK9 pKa = 9.85CFLLFHH15 pKa = 7.34IINFLGTYY23 pKa = 9.08NVGWVPGTPLCAAQAVDD40 pKa = 5.09GITSRR45 pKa = 11.84EE46 pKa = 3.86MEE48 pKa = 4.35INATLAPSSGATLSLLVTLSNNNPPTAMRR77 pKa = 11.84PPVAQNGEE85 pKa = 4.4SVSIDD90 pKa = 3.44NSSAPASDD98 pKa = 3.58PTTSNPSSPEE108 pKa = 3.89EE109 pKa = 4.34APTAAPITPIPTSTAAQSAEE129 pKa = 4.16HH130 pKa = 6.57VGEE133 pKa = 4.3TDD135 pKa = 3.67SEE137 pKa = 4.7APTPLPTTPKK147 pKa = 10.05PSSQEE152 pKa = 4.07DD153 pKa = 4.65EE154 pKa = 4.26PTMTSPTEE162 pKa = 4.1TPPTTTAAISTEE174 pKa = 3.96QDD176 pKa = 3.37DD177 pKa = 4.15EE178 pKa = 4.67TEE180 pKa = 4.59PEE182 pKa = 4.52SPAPPPATPEE192 pKa = 4.3PEE194 pKa = 4.13TTTPTKK200 pKa = 10.55NQEE203 pKa = 4.1DD204 pKa = 4.74EE205 pKa = 4.28PTINTSDD212 pKa = 3.43QGDD215 pKa = 4.09DD216 pKa = 3.54SSSDD220 pKa = 3.34IPAGTPGPTTPPKK233 pKa = 10.56QEE235 pKa = 4.03TEE237 pKa = 3.93TTKK240 pKa = 10.82PVDD243 pKa = 3.5SKK245 pKa = 11.4PQVEE249 pKa = 4.78PNDD252 pKa = 4.07SAPSDD257 pKa = 3.54IPEE260 pKa = 4.6TSDD263 pKa = 3.31STPTPVTDD271 pKa = 3.57PTSPPSVEE279 pKa = 4.07EE280 pKa = 3.94TSPAEE285 pKa = 4.02PSTPDD290 pKa = 3.23STPPADD296 pKa = 4.22PPAPQPTPPAEE307 pKa = 4.41PSTPDD312 pKa = 3.32STPQDD317 pKa = 3.73EE318 pKa = 4.92PSTPDD323 pKa = 3.14STPPADD329 pKa = 4.22PPAPQPTPPAEE340 pKa = 4.36PSTPEE345 pKa = 3.86STPPADD351 pKa = 4.1PPAAQPTPQAEE362 pKa = 4.3PSTPDD367 pKa = 2.98STSPSDD373 pKa = 3.85PPASQPTLPEE383 pKa = 4.52KK384 pKa = 10.49PPSPEE389 pKa = 3.55PMTPQSGPAITPEE402 pKa = 3.62IATPSTTEE410 pKa = 3.65PGAGKK415 pKa = 9.61DD416 pKa = 3.31ASMGATTTQAASALTTKK433 pKa = 10.24PMRR436 pKa = 11.84VVPDD440 pKa = 3.58VSTMLWIRR448 pKa = 11.84PTVAIVLIFLLMSIFHH464 pKa = 6.88IMYY467 pKa = 8.91CVCLHH472 pKa = 6.12EE473 pKa = 4.44
MM1 pKa = 7.57CGVKK5 pKa = 10.28PIVKK9 pKa = 9.85CFLLFHH15 pKa = 7.34IINFLGTYY23 pKa = 9.08NVGWVPGTPLCAAQAVDD40 pKa = 5.09GITSRR45 pKa = 11.84EE46 pKa = 3.86MEE48 pKa = 4.35INATLAPSSGATLSLLVTLSNNNPPTAMRR77 pKa = 11.84PPVAQNGEE85 pKa = 4.4SVSIDD90 pKa = 3.44NSSAPASDD98 pKa = 3.58PTTSNPSSPEE108 pKa = 3.89EE109 pKa = 4.34APTAAPITPIPTSTAAQSAEE129 pKa = 4.16HH130 pKa = 6.57VGEE133 pKa = 4.3TDD135 pKa = 3.67SEE137 pKa = 4.7APTPLPTTPKK147 pKa = 10.05PSSQEE152 pKa = 4.07DD153 pKa = 4.65EE154 pKa = 4.26PTMTSPTEE162 pKa = 4.1TPPTTTAAISTEE174 pKa = 3.96QDD176 pKa = 3.37DD177 pKa = 4.15EE178 pKa = 4.67TEE180 pKa = 4.59PEE182 pKa = 4.52SPAPPPATPEE192 pKa = 4.3PEE194 pKa = 4.13TTTPTKK200 pKa = 10.55NQEE203 pKa = 4.1DD204 pKa = 4.74EE205 pKa = 4.28PTINTSDD212 pKa = 3.43QGDD215 pKa = 4.09DD216 pKa = 3.54SSSDD220 pKa = 3.34IPAGTPGPTTPPKK233 pKa = 10.56QEE235 pKa = 4.03TEE237 pKa = 3.93TTKK240 pKa = 10.82PVDD243 pKa = 3.5SKK245 pKa = 11.4PQVEE249 pKa = 4.78PNDD252 pKa = 4.07SAPSDD257 pKa = 3.54IPEE260 pKa = 4.6TSDD263 pKa = 3.31STPTPVTDD271 pKa = 3.57PTSPPSVEE279 pKa = 4.07EE280 pKa = 3.94TSPAEE285 pKa = 4.02PSTPDD290 pKa = 3.23STPPADD296 pKa = 4.22PPAPQPTPPAEE307 pKa = 4.41PSTPDD312 pKa = 3.32STPQDD317 pKa = 3.73EE318 pKa = 4.92PSTPDD323 pKa = 3.14STPPADD329 pKa = 4.22PPAPQPTPPAEE340 pKa = 4.36PSTPEE345 pKa = 3.86STPPADD351 pKa = 4.1PPAAQPTPQAEE362 pKa = 4.3PSTPDD367 pKa = 2.98STSPSDD373 pKa = 3.85PPASQPTLPEE383 pKa = 4.52KK384 pKa = 10.49PPSPEE389 pKa = 3.55PMTPQSGPAITPEE402 pKa = 3.62IATPSTTEE410 pKa = 3.65PGAGKK415 pKa = 9.61DD416 pKa = 3.31ASMGATTTQAASALTTKK433 pKa = 10.24PMRR436 pKa = 11.84VVPDD440 pKa = 3.58VSTMLWIRR448 pKa = 11.84PTVAIVLIFLLMSIFHH464 pKa = 6.88IMYY467 pKa = 8.91CVCLHH472 pKa = 6.12EE473 pKa = 4.44
Molecular weight: 48.6 kDa
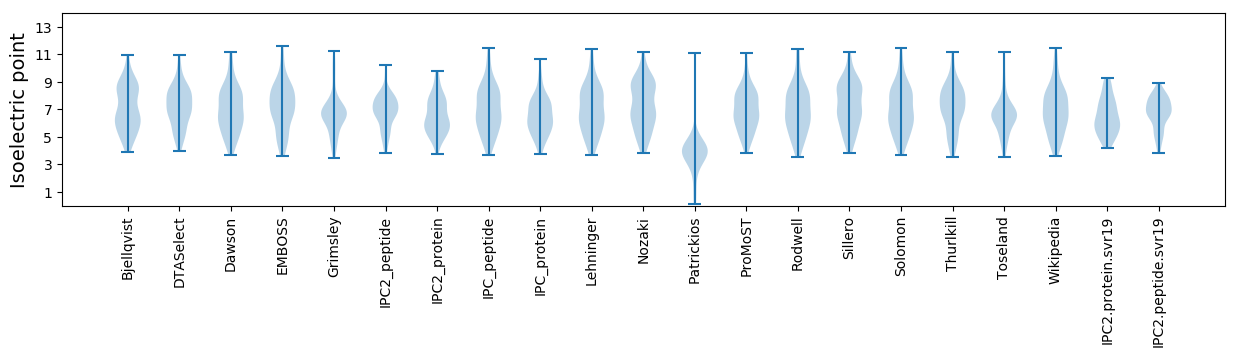
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0U1L1|D0U1L1_9GAMA Thymidine kinase OS=Wood mouse herpesvirus OX=432370 GN=ORF21 PE=3 SV=1
MM1 pKa = 7.94DD2 pKa = 4.34EE3 pKa = 4.88KK4 pKa = 11.09GAKK7 pKa = 9.11QLLNKK12 pKa = 9.92LPKK15 pKa = 9.66RR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84AGKK22 pKa = 8.89LAHH25 pKa = 5.4VRR27 pKa = 11.84CYY29 pKa = 10.57RR30 pKa = 11.84AMQGASNILQLLEE43 pKa = 4.08SLKK46 pKa = 10.16IAHH49 pKa = 6.52QITHH53 pKa = 6.65PVGSRR58 pKa = 11.84LFFEE62 pKa = 4.5VTLGRR67 pKa = 11.84RR68 pKa = 11.84VVDD71 pKa = 4.63AIIVVFSEE79 pKa = 4.29SSPHH83 pKa = 4.79IHH85 pKa = 6.93CFLIEE90 pKa = 4.49LKK92 pKa = 9.19TCKK95 pKa = 10.06INFFNHH101 pKa = 5.93NSTTRR106 pKa = 11.84EE107 pKa = 3.83AQRR110 pKa = 11.84VEE112 pKa = 4.25GSSQLRR118 pKa = 11.84DD119 pKa = 3.24SARR122 pKa = 11.84ALTLLAPVGTDD133 pKa = 3.15PCRR136 pKa = 11.84VTAHH140 pKa = 7.4LIFKK144 pKa = 7.91SQRR147 pKa = 11.84GLHH150 pKa = 5.06TLKK153 pKa = 10.72SYY155 pKa = 9.43TLTWMTHH162 pKa = 5.79TINTQKK168 pKa = 10.68VALLNFLGLNADD180 pKa = 3.77NEE182 pKa = 4.23LRR184 pKa = 11.84ACLTKK189 pKa = 10.59GLPPARR195 pKa = 11.84SLGRR199 pKa = 11.84RR200 pKa = 11.84SHH202 pKa = 6.01VCLLEE207 pKa = 4.24PKK209 pKa = 9.12PQKK212 pKa = 9.84HH213 pKa = 5.55LKK215 pKa = 8.78NRR217 pKa = 11.84RR218 pKa = 11.84GGASRR223 pKa = 11.84NQKK226 pKa = 9.72ARR228 pKa = 11.84RR229 pKa = 11.84QGVGPKK235 pKa = 9.8VSHH238 pKa = 7.42DD239 pKa = 3.45KK240 pKa = 10.59TGNAPAHH247 pKa = 5.52AAGRR251 pKa = 11.84AGG253 pKa = 3.44
MM1 pKa = 7.94DD2 pKa = 4.34EE3 pKa = 4.88KK4 pKa = 11.09GAKK7 pKa = 9.11QLLNKK12 pKa = 9.92LPKK15 pKa = 9.66RR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84AGKK22 pKa = 8.89LAHH25 pKa = 5.4VRR27 pKa = 11.84CYY29 pKa = 10.57RR30 pKa = 11.84AMQGASNILQLLEE43 pKa = 4.08SLKK46 pKa = 10.16IAHH49 pKa = 6.52QITHH53 pKa = 6.65PVGSRR58 pKa = 11.84LFFEE62 pKa = 4.5VTLGRR67 pKa = 11.84RR68 pKa = 11.84VVDD71 pKa = 4.63AIIVVFSEE79 pKa = 4.29SSPHH83 pKa = 4.79IHH85 pKa = 6.93CFLIEE90 pKa = 4.49LKK92 pKa = 9.19TCKK95 pKa = 10.06INFFNHH101 pKa = 5.93NSTTRR106 pKa = 11.84EE107 pKa = 3.83AQRR110 pKa = 11.84VEE112 pKa = 4.25GSSQLRR118 pKa = 11.84DD119 pKa = 3.24SARR122 pKa = 11.84ALTLLAPVGTDD133 pKa = 3.15PCRR136 pKa = 11.84VTAHH140 pKa = 7.4LIFKK144 pKa = 7.91SQRR147 pKa = 11.84GLHH150 pKa = 5.06TLKK153 pKa = 10.72SYY155 pKa = 9.43TLTWMTHH162 pKa = 5.79TINTQKK168 pKa = 10.68VALLNFLGLNADD180 pKa = 3.77NEE182 pKa = 4.23LRR184 pKa = 11.84ACLTKK189 pKa = 10.59GLPPARR195 pKa = 11.84SLGRR199 pKa = 11.84RR200 pKa = 11.84SHH202 pKa = 6.01VCLLEE207 pKa = 4.24PKK209 pKa = 9.12PQKK212 pKa = 9.84HH213 pKa = 5.55LKK215 pKa = 8.78NRR217 pKa = 11.84RR218 pKa = 11.84GGASRR223 pKa = 11.84NQKK226 pKa = 9.72ARR228 pKa = 11.84RR229 pKa = 11.84QGVGPKK235 pKa = 9.8VSHH238 pKa = 7.42DD239 pKa = 3.45KK240 pKa = 10.59TGNAPAHH247 pKa = 5.52AAGRR251 pKa = 11.84AGG253 pKa = 3.44
Molecular weight: 28.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
35050 |
75 |
2455 |
473.6 |
53.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.886 ± 0.178 | 2.636 ± 0.175 |
4.805 ± 0.144 | 5.193 ± 0.138 |
4.514 ± 0.168 | 5.101 ± 0.207 |
2.862 ± 0.112 | 6.117 ± 0.179 |
5.187 ± 0.228 | 10.439 ± 0.268 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.491 ± 0.119 | 4.362 ± 0.119 |
6.191 ± 0.338 | 3.74 ± 0.12 |
4.217 ± 0.179 | 8.18 ± 0.201 |
7.384 ± 0.253 | 6.163 ± 0.204 |
1.096 ± 0.066 | 3.438 ± 0.13 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
