
HCBI8.215 virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus bovas1; Bovine associated gemykibivirus 1
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
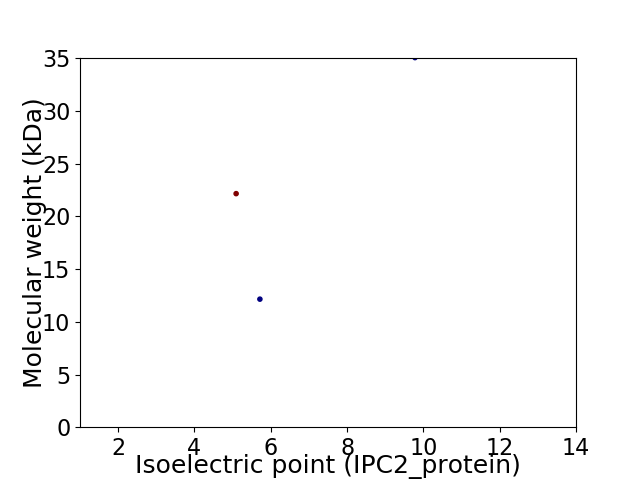
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
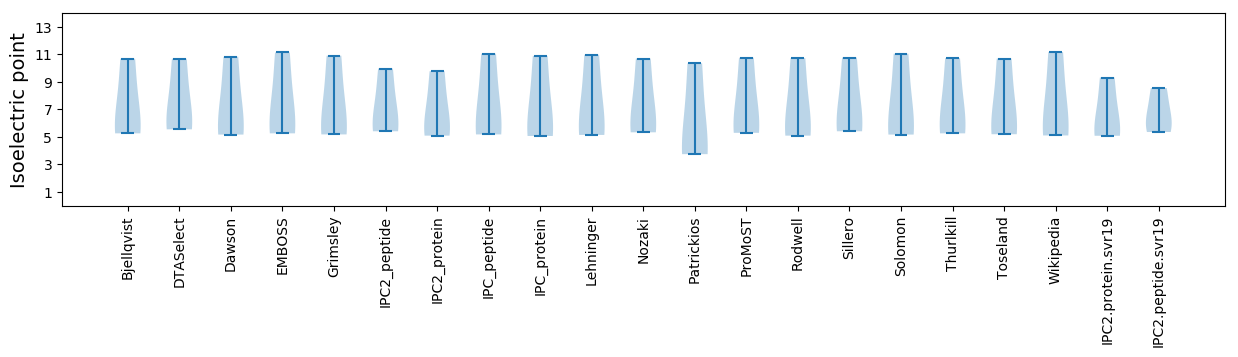
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077XLY1|A0A077XLY1_9VIRU Replication associated protein OS=HCBI8.215 virus OX=1516079 GN=Rep PE=3 SV=1
MM1 pKa = 6.81STFRR5 pKa = 11.84FYY7 pKa = 11.55ARR9 pKa = 11.84YY10 pKa = 9.84ALLTYY15 pKa = 8.76AQCGGLDD22 pKa = 3.42PFTIVSHH29 pKa = 6.94IGDD32 pKa = 4.26LGGEE36 pKa = 4.27CIIGRR41 pKa = 11.84EE42 pKa = 4.08AHH44 pKa = 6.42SDD46 pKa = 3.43GGTHH50 pKa = 6.34LHH52 pKa = 6.56AFVDD56 pKa = 4.72FGRR59 pKa = 11.84RR60 pKa = 11.84FQSRR64 pKa = 11.84RR65 pKa = 11.84AAVFDD70 pKa = 3.38VGGFHH75 pKa = 7.23PNISITKK82 pKa = 9.17RR83 pKa = 11.84DD84 pKa = 3.72PQVHH88 pKa = 6.21YY89 pKa = 10.59DD90 pKa = 3.67YY91 pKa = 10.85AIKK94 pKa = 10.96DD95 pKa = 3.58GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.07RR105 pKa = 11.84PAADD109 pKa = 3.1ASAGVEE115 pKa = 4.2NKK117 pKa = 9.24HH118 pKa = 5.67HH119 pKa = 6.42QIVLAPTRR127 pKa = 11.84AEE129 pKa = 3.7FFEE132 pKa = 4.3AVRR135 pKa = 11.84DD136 pKa = 3.68LDD138 pKa = 3.68PRR140 pKa = 11.84LLLTSFPSLEE150 pKa = 4.79KK151 pKa = 10.83YY152 pKa = 10.75ADD154 pKa = 2.59WHH156 pKa = 6.06YY157 pKa = 11.17RR158 pKa = 11.84VDD160 pKa = 4.05PDD162 pKa = 4.5PYY164 pKa = 10.56RR165 pKa = 11.84HH166 pKa = 6.22DD167 pKa = 3.79PEE169 pKa = 5.17YY170 pKa = 11.05EE171 pKa = 3.74FDD173 pKa = 3.34TSQFPEE179 pKa = 4.12LNQWAEE185 pKa = 4.86DD186 pKa = 4.09YY187 pKa = 10.71IGWDD191 pKa = 3.05ATVRR195 pKa = 11.84GG196 pKa = 4.08
MM1 pKa = 6.81STFRR5 pKa = 11.84FYY7 pKa = 11.55ARR9 pKa = 11.84YY10 pKa = 9.84ALLTYY15 pKa = 8.76AQCGGLDD22 pKa = 3.42PFTIVSHH29 pKa = 6.94IGDD32 pKa = 4.26LGGEE36 pKa = 4.27CIIGRR41 pKa = 11.84EE42 pKa = 4.08AHH44 pKa = 6.42SDD46 pKa = 3.43GGTHH50 pKa = 6.34LHH52 pKa = 6.56AFVDD56 pKa = 4.72FGRR59 pKa = 11.84RR60 pKa = 11.84FQSRR64 pKa = 11.84RR65 pKa = 11.84AAVFDD70 pKa = 3.38VGGFHH75 pKa = 7.23PNISITKK82 pKa = 9.17RR83 pKa = 11.84DD84 pKa = 3.72PQVHH88 pKa = 6.21YY89 pKa = 10.59DD90 pKa = 3.67YY91 pKa = 10.85AIKK94 pKa = 10.96DD95 pKa = 3.58GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.07RR105 pKa = 11.84PAADD109 pKa = 3.1ASAGVEE115 pKa = 4.2NKK117 pKa = 9.24HH118 pKa = 5.67HH119 pKa = 6.42QIVLAPTRR127 pKa = 11.84AEE129 pKa = 3.7FFEE132 pKa = 4.3AVRR135 pKa = 11.84DD136 pKa = 3.68LDD138 pKa = 3.68PRR140 pKa = 11.84LLLTSFPSLEE150 pKa = 4.79KK151 pKa = 10.83YY152 pKa = 10.75ADD154 pKa = 2.59WHH156 pKa = 6.06YY157 pKa = 11.17RR158 pKa = 11.84VDD160 pKa = 4.05PDD162 pKa = 4.5PYY164 pKa = 10.56RR165 pKa = 11.84HH166 pKa = 6.22DD167 pKa = 3.79PEE169 pKa = 5.17YY170 pKa = 11.05EE171 pKa = 3.74FDD173 pKa = 3.34TSQFPEE179 pKa = 4.12LNQWAEE185 pKa = 4.86DD186 pKa = 4.09YY187 pKa = 10.71IGWDD191 pKa = 3.05ATVRR195 pKa = 11.84GG196 pKa = 4.08
Molecular weight: 22.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077XLY1|A0A077XLY1_9VIRU Replication associated protein OS=HCBI8.215 virus OX=1516079 GN=Rep PE=3 SV=1
MM1 pKa = 7.76AFRR4 pKa = 11.84NRR6 pKa = 11.84RR7 pKa = 11.84FQAGRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FTPRR19 pKa = 11.84RR20 pKa = 11.84STFRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SFRR29 pKa = 11.84NGRR32 pKa = 11.84SRR34 pKa = 11.84TTRR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 9.42SRR41 pKa = 11.84PRR43 pKa = 11.84TNVRR47 pKa = 11.84RR48 pKa = 11.84VRR50 pKa = 11.84NLASRR55 pKa = 11.84KK56 pKa = 9.58CRR58 pKa = 11.84DD59 pKa = 3.3NMICTPIDD67 pKa = 3.5DD68 pKa = 4.45TGTPQTPGPVSMDD81 pKa = 3.02ASSVYY86 pKa = 10.76GFIFSPTARR95 pKa = 11.84TAGTQLEE102 pKa = 4.34SGEE105 pKa = 4.35RR106 pKa = 11.84VNAPSSAQRR115 pKa = 11.84RR116 pKa = 11.84KK117 pKa = 9.22SKK119 pKa = 10.67CYY121 pKa = 8.68VRR123 pKa = 11.84GYY125 pKa = 10.86GEE127 pKa = 4.14TVEE130 pKa = 4.56IQTDD134 pKa = 3.94DD135 pKa = 3.29ASPWVWRR142 pKa = 11.84RR143 pKa = 11.84IVFSTIGLASQFDD156 pKa = 3.87PGLLYY161 pKa = 11.27NNDD164 pKa = 3.46EE165 pKa = 4.1TRR167 pKa = 11.84GWGRR171 pKa = 11.84SLFNIRR177 pKa = 11.84SGTQQASILEE187 pKa = 4.34GVVFRR192 pKa = 11.84YY193 pKa = 9.89LFQGTEE199 pKa = 4.11GVDD202 pKa = 3.09WANVMTATTAPRR214 pKa = 11.84HH215 pKa = 4.66VRR217 pKa = 11.84VYY219 pKa = 9.64SDD221 pKa = 3.29RR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84VIQSPNSEE232 pKa = 3.9GGTIRR237 pKa = 11.84LSKK240 pKa = 10.56HH241 pKa = 5.14YY242 pKa = 10.64DD243 pKa = 3.66GINRR247 pKa = 11.84SIIYY251 pKa = 10.32DD252 pKa = 3.65DD253 pKa = 4.14VEE255 pKa = 4.71IGAGAKK261 pKa = 7.94ATNAIASGTPYY272 pKa = 11.37GNMGDD277 pKa = 4.75LFVLDD282 pKa = 4.95FFSSASDD289 pKa = 4.03DD290 pKa = 3.95EE291 pKa = 4.57TSGASFQPHH300 pKa = 5.72GKK302 pKa = 9.87FYY304 pKa = 9.48WHH306 pKa = 7.28EE307 pKa = 4.29GQSAA311 pKa = 3.13
MM1 pKa = 7.76AFRR4 pKa = 11.84NRR6 pKa = 11.84RR7 pKa = 11.84FQAGRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FTPRR19 pKa = 11.84RR20 pKa = 11.84STFRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SFRR29 pKa = 11.84NGRR32 pKa = 11.84SRR34 pKa = 11.84TTRR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 9.42SRR41 pKa = 11.84PRR43 pKa = 11.84TNVRR47 pKa = 11.84RR48 pKa = 11.84VRR50 pKa = 11.84NLASRR55 pKa = 11.84KK56 pKa = 9.58CRR58 pKa = 11.84DD59 pKa = 3.3NMICTPIDD67 pKa = 3.5DD68 pKa = 4.45TGTPQTPGPVSMDD81 pKa = 3.02ASSVYY86 pKa = 10.76GFIFSPTARR95 pKa = 11.84TAGTQLEE102 pKa = 4.34SGEE105 pKa = 4.35RR106 pKa = 11.84VNAPSSAQRR115 pKa = 11.84RR116 pKa = 11.84KK117 pKa = 9.22SKK119 pKa = 10.67CYY121 pKa = 8.68VRR123 pKa = 11.84GYY125 pKa = 10.86GEE127 pKa = 4.14TVEE130 pKa = 4.56IQTDD134 pKa = 3.94DD135 pKa = 3.29ASPWVWRR142 pKa = 11.84RR143 pKa = 11.84IVFSTIGLASQFDD156 pKa = 3.87PGLLYY161 pKa = 11.27NNDD164 pKa = 3.46EE165 pKa = 4.1TRR167 pKa = 11.84GWGRR171 pKa = 11.84SLFNIRR177 pKa = 11.84SGTQQASILEE187 pKa = 4.34GVVFRR192 pKa = 11.84YY193 pKa = 9.89LFQGTEE199 pKa = 4.11GVDD202 pKa = 3.09WANVMTATTAPRR214 pKa = 11.84HH215 pKa = 4.66VRR217 pKa = 11.84VYY219 pKa = 9.64SDD221 pKa = 3.29RR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84VIQSPNSEE232 pKa = 3.9GGTIRR237 pKa = 11.84LSKK240 pKa = 10.56HH241 pKa = 5.14YY242 pKa = 10.64DD243 pKa = 3.66GINRR247 pKa = 11.84SIIYY251 pKa = 10.32DD252 pKa = 3.65DD253 pKa = 4.14VEE255 pKa = 4.71IGAGAKK261 pKa = 7.94ATNAIASGTPYY272 pKa = 11.37GNMGDD277 pKa = 4.75LFVLDD282 pKa = 4.95FFSSASDD289 pKa = 4.03DD290 pKa = 3.95EE291 pKa = 4.57TSGASFQPHH300 pKa = 5.72GKK302 pKa = 9.87FYY304 pKa = 9.48WHH306 pKa = 7.28EE307 pKa = 4.29GQSAA311 pKa = 3.13
Molecular weight: 35.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
611 |
104 |
311 |
203.7 |
23.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.692 ± 0.924 | 1.146 ± 0.206 |
7.201 ± 1.186 | 4.583 ± 0.643 |
6.056 ± 0.363 | 8.511 ± 0.476 |
2.455 ± 1.168 | 4.91 ± 0.242 |
3.601 ± 2.109 | 5.237 ± 1.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.309 ± 0.359 | 3.273 ± 0.834 |
4.746 ± 0.406 | 3.437 ± 0.259 |
9.493 ± 2.709 | 7.692 ± 1.514 |
6.219 ± 1.119 | 5.728 ± 0.268 |
2.128 ± 0.714 | 4.583 ± 0.696 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
