
Lye Green virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
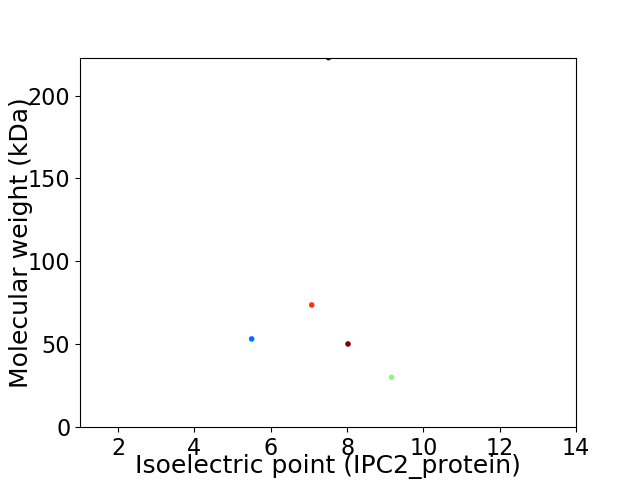
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140HEQ6|A0A140HEQ6_9RHAB VP2 OS=Lye Green virus OX=1807808 PE=4 SV=1
MM1 pKa = 7.18YY2 pKa = 8.91PNHH5 pKa = 6.65HH6 pKa = 6.48NLRR9 pKa = 11.84GSGQHH14 pKa = 7.32DD15 pKa = 4.47DD16 pKa = 3.5IQFCEE21 pKa = 4.56GNAEE25 pKa = 4.01VLNYY29 pKa = 10.03LKK31 pKa = 10.81NVTPDD36 pKa = 3.34MDD38 pKa = 4.76NDD40 pKa = 4.13TPPLDD45 pKa = 3.4WTEE48 pKa = 3.88EE49 pKa = 4.35TFPSMAMGSKK59 pKa = 9.84IKK61 pKa = 9.47TRR63 pKa = 11.84PSILDD68 pKa = 3.29QVNAASIVLNWYY80 pKa = 9.55VSGIQRR86 pKa = 11.84RR87 pKa = 11.84ILVPIEE93 pKa = 3.84TVIVGMTTSGDD104 pKa = 3.34ANYY107 pKa = 10.33VFGDD111 pKa = 4.07EE112 pKa = 5.05IFDD115 pKa = 3.52ATTGIEE121 pKa = 4.68LEE123 pKa = 4.13PQPAVEE129 pKa = 4.44MLTPSEE135 pKa = 4.16LEE137 pKa = 3.59RR138 pKa = 11.84WRR140 pKa = 11.84AAAIGAGPEE149 pKa = 4.17LPPMNPMGGIPIAQGDD165 pKa = 3.9ASKK168 pKa = 10.55LASAVPDD175 pKa = 3.35TTLWYY180 pKa = 10.77KK181 pKa = 11.02KK182 pKa = 8.05MWEE185 pKa = 4.11LLHH188 pKa = 5.96VVVVADD194 pKa = 3.55SAEE197 pKa = 4.32TAEE200 pKa = 4.14QTAGRR205 pKa = 11.84RR206 pKa = 11.84YY207 pKa = 8.21ALNYY211 pKa = 9.25IGYY214 pKa = 8.69ICLLVIRR221 pKa = 11.84GVAKK225 pKa = 10.58NHH227 pKa = 5.61ATVIAALEE235 pKa = 4.17KK236 pKa = 10.5NLPDD240 pKa = 3.35RR241 pKa = 11.84VTAFWPYY248 pKa = 10.73DD249 pKa = 3.24HH250 pKa = 7.19TMPKK254 pKa = 10.29PPMGHH259 pKa = 7.27DD260 pKa = 2.9ISIRR264 pKa = 11.84DD265 pKa = 3.43IEE267 pKa = 4.31NALEE271 pKa = 4.3RR272 pKa = 11.84GSSHH276 pKa = 5.57TRR278 pKa = 11.84RR279 pKa = 11.84LLVNLIQSSLTTEE292 pKa = 3.74PTKK295 pKa = 10.71RR296 pKa = 11.84GILMSGCMKK305 pKa = 10.53SLQDD309 pKa = 3.45SGLGALHH316 pKa = 6.45WVFEE320 pKa = 4.22ASRR323 pKa = 11.84AIEE326 pKa = 4.07VEE328 pKa = 3.77PRR330 pKa = 11.84ALMEE334 pKa = 4.22AMFYY338 pKa = 11.16GPFIGSTMVVGRR350 pKa = 11.84FLSEE354 pKa = 3.98AKK356 pKa = 10.33NSSRR360 pKa = 11.84SWKK363 pKa = 7.89WARR366 pKa = 11.84VLNDD370 pKa = 3.94GYY372 pKa = 10.14LTALSVKK379 pKa = 10.06NNKK382 pKa = 9.53IYY384 pKa = 10.92VATLVALVAEE394 pKa = 5.3LKK396 pKa = 10.02PDD398 pKa = 3.52HH399 pKa = 7.05AIWNIPSLALSNWMKK414 pKa = 10.6HH415 pKa = 5.05RR416 pKa = 11.84ALEE419 pKa = 4.21HH420 pKa = 5.86AQSISAEE427 pKa = 4.3LYY429 pKa = 10.29DD430 pKa = 4.34AAHH433 pKa = 6.32FQAGSTSGKK442 pKa = 10.02NVIARR447 pKa = 11.84IRR449 pKa = 11.84LAKK452 pKa = 10.5ANARR456 pKa = 11.84LTTIPEE462 pKa = 4.38ANPLPNVQAQMMDD475 pKa = 3.48DD476 pKa = 3.65QEE478 pKa = 5.06LGEE481 pKa = 4.49VII483 pKa = 4.57
MM1 pKa = 7.18YY2 pKa = 8.91PNHH5 pKa = 6.65HH6 pKa = 6.48NLRR9 pKa = 11.84GSGQHH14 pKa = 7.32DD15 pKa = 4.47DD16 pKa = 3.5IQFCEE21 pKa = 4.56GNAEE25 pKa = 4.01VLNYY29 pKa = 10.03LKK31 pKa = 10.81NVTPDD36 pKa = 3.34MDD38 pKa = 4.76NDD40 pKa = 4.13TPPLDD45 pKa = 3.4WTEE48 pKa = 3.88EE49 pKa = 4.35TFPSMAMGSKK59 pKa = 9.84IKK61 pKa = 9.47TRR63 pKa = 11.84PSILDD68 pKa = 3.29QVNAASIVLNWYY80 pKa = 9.55VSGIQRR86 pKa = 11.84RR87 pKa = 11.84ILVPIEE93 pKa = 3.84TVIVGMTTSGDD104 pKa = 3.34ANYY107 pKa = 10.33VFGDD111 pKa = 4.07EE112 pKa = 5.05IFDD115 pKa = 3.52ATTGIEE121 pKa = 4.68LEE123 pKa = 4.13PQPAVEE129 pKa = 4.44MLTPSEE135 pKa = 4.16LEE137 pKa = 3.59RR138 pKa = 11.84WRR140 pKa = 11.84AAAIGAGPEE149 pKa = 4.17LPPMNPMGGIPIAQGDD165 pKa = 3.9ASKK168 pKa = 10.55LASAVPDD175 pKa = 3.35TTLWYY180 pKa = 10.77KK181 pKa = 11.02KK182 pKa = 8.05MWEE185 pKa = 4.11LLHH188 pKa = 5.96VVVVADD194 pKa = 3.55SAEE197 pKa = 4.32TAEE200 pKa = 4.14QTAGRR205 pKa = 11.84RR206 pKa = 11.84YY207 pKa = 8.21ALNYY211 pKa = 9.25IGYY214 pKa = 8.69ICLLVIRR221 pKa = 11.84GVAKK225 pKa = 10.58NHH227 pKa = 5.61ATVIAALEE235 pKa = 4.17KK236 pKa = 10.5NLPDD240 pKa = 3.35RR241 pKa = 11.84VTAFWPYY248 pKa = 10.73DD249 pKa = 3.24HH250 pKa = 7.19TMPKK254 pKa = 10.29PPMGHH259 pKa = 7.27DD260 pKa = 2.9ISIRR264 pKa = 11.84DD265 pKa = 3.43IEE267 pKa = 4.31NALEE271 pKa = 4.3RR272 pKa = 11.84GSSHH276 pKa = 5.57TRR278 pKa = 11.84RR279 pKa = 11.84LLVNLIQSSLTTEE292 pKa = 3.74PTKK295 pKa = 10.71RR296 pKa = 11.84GILMSGCMKK305 pKa = 10.53SLQDD309 pKa = 3.45SGLGALHH316 pKa = 6.45WVFEE320 pKa = 4.22ASRR323 pKa = 11.84AIEE326 pKa = 4.07VEE328 pKa = 3.77PRR330 pKa = 11.84ALMEE334 pKa = 4.22AMFYY338 pKa = 11.16GPFIGSTMVVGRR350 pKa = 11.84FLSEE354 pKa = 3.98AKK356 pKa = 10.33NSSRR360 pKa = 11.84SWKK363 pKa = 7.89WARR366 pKa = 11.84VLNDD370 pKa = 3.94GYY372 pKa = 10.14LTALSVKK379 pKa = 10.06NNKK382 pKa = 9.53IYY384 pKa = 10.92VATLVALVAEE394 pKa = 5.3LKK396 pKa = 10.02PDD398 pKa = 3.52HH399 pKa = 7.05AIWNIPSLALSNWMKK414 pKa = 10.6HH415 pKa = 5.05RR416 pKa = 11.84ALEE419 pKa = 4.21HH420 pKa = 5.86AQSISAEE427 pKa = 4.3LYY429 pKa = 10.29DD430 pKa = 4.34AAHH433 pKa = 6.32FQAGSTSGKK442 pKa = 10.02NVIARR447 pKa = 11.84IRR449 pKa = 11.84LAKK452 pKa = 10.5ANARR456 pKa = 11.84LTTIPEE462 pKa = 4.38ANPLPNVQAQMMDD475 pKa = 3.48DD476 pKa = 3.65QEE478 pKa = 5.06LGEE481 pKa = 4.49VII483 pKa = 4.57
Molecular weight: 53.25 kDa
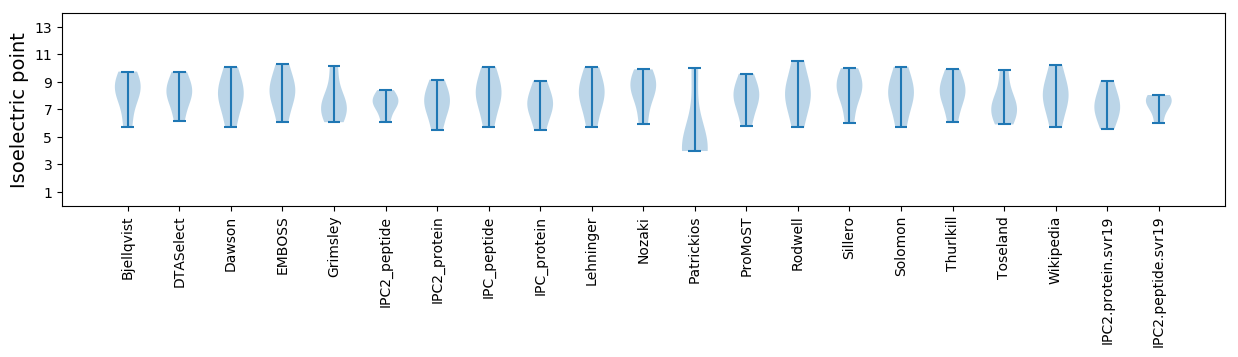
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140HEQ8|A0A140HEQ8_9RHAB Putative glycoprotein OS=Lye Green virus OX=1807808 PE=4 SV=1
MM1 pKa = 7.4NLLPGASIHH10 pKa = 6.02QRR12 pKa = 11.84DD13 pKa = 3.9KK14 pKa = 10.75ISRR17 pKa = 11.84FRR19 pKa = 11.84KK20 pKa = 9.44NVLHH24 pKa = 6.79LEE26 pKa = 4.16KK27 pKa = 10.72VVNKK31 pKa = 9.75VFTSYY36 pKa = 10.72TITFVKK42 pKa = 8.78PTSNMSSRR50 pKa = 11.84LIPRR54 pKa = 11.84IPVTKK59 pKa = 10.46SPLKK63 pKa = 9.63TAIQVEE69 pKa = 4.23LRR71 pKa = 11.84LWGAQGLDD79 pKa = 3.07NTLLKK84 pKa = 10.14KK85 pKa = 10.36ICVCVIWEE93 pKa = 4.31SYY95 pKa = 10.18KK96 pKa = 11.15GVVDD100 pKa = 5.86DD101 pKa = 4.85PEE103 pKa = 4.44LWSHH107 pKa = 7.05ASQALYY113 pKa = 10.8QRR115 pKa = 11.84MKK117 pKa = 10.47RR118 pKa = 11.84KK119 pKa = 10.26CEE121 pKa = 3.55WAPIMSQSSAQFLVSTTWFSLHH143 pKa = 6.47PRR145 pKa = 11.84NLYY148 pKa = 10.43EE149 pKa = 4.5GFSQKK154 pKa = 8.89RR155 pKa = 11.84TFSRR159 pKa = 11.84LFHH162 pKa = 6.01FHH164 pKa = 6.88SPTGGILSVDD174 pKa = 3.38YY175 pKa = 11.25VITLRR180 pKa = 11.84HH181 pKa = 5.25NTWDD185 pKa = 3.3LKK187 pKa = 10.94EE188 pKa = 4.05YY189 pKa = 11.1GNMVLMGYY197 pKa = 10.2LCLEE201 pKa = 4.02QTNYY205 pKa = 9.87VPNPKK210 pKa = 10.2GVISLAIEE218 pKa = 4.16DD219 pKa = 4.84LKK221 pKa = 11.23PLINFDD227 pKa = 3.83PYY229 pKa = 11.62QMVRR233 pKa = 11.84LLGLSPMIEE242 pKa = 4.4DD243 pKa = 6.03PIPAPSKK250 pKa = 9.58TRR252 pKa = 11.84SILPAKK258 pKa = 9.99FFKK261 pKa = 10.75KK262 pKa = 10.49
MM1 pKa = 7.4NLLPGASIHH10 pKa = 6.02QRR12 pKa = 11.84DD13 pKa = 3.9KK14 pKa = 10.75ISRR17 pKa = 11.84FRR19 pKa = 11.84KK20 pKa = 9.44NVLHH24 pKa = 6.79LEE26 pKa = 4.16KK27 pKa = 10.72VVNKK31 pKa = 9.75VFTSYY36 pKa = 10.72TITFVKK42 pKa = 8.78PTSNMSSRR50 pKa = 11.84LIPRR54 pKa = 11.84IPVTKK59 pKa = 10.46SPLKK63 pKa = 9.63TAIQVEE69 pKa = 4.23LRR71 pKa = 11.84LWGAQGLDD79 pKa = 3.07NTLLKK84 pKa = 10.14KK85 pKa = 10.36ICVCVIWEE93 pKa = 4.31SYY95 pKa = 10.18KK96 pKa = 11.15GVVDD100 pKa = 5.86DD101 pKa = 4.85PEE103 pKa = 4.44LWSHH107 pKa = 7.05ASQALYY113 pKa = 10.8QRR115 pKa = 11.84MKK117 pKa = 10.47RR118 pKa = 11.84KK119 pKa = 10.26CEE121 pKa = 3.55WAPIMSQSSAQFLVSTTWFSLHH143 pKa = 6.47PRR145 pKa = 11.84NLYY148 pKa = 10.43EE149 pKa = 4.5GFSQKK154 pKa = 8.89RR155 pKa = 11.84TFSRR159 pKa = 11.84LFHH162 pKa = 6.01FHH164 pKa = 6.88SPTGGILSVDD174 pKa = 3.38YY175 pKa = 11.25VITLRR180 pKa = 11.84HH181 pKa = 5.25NTWDD185 pKa = 3.3LKK187 pKa = 10.94EE188 pKa = 4.05YY189 pKa = 11.1GNMVLMGYY197 pKa = 10.2LCLEE201 pKa = 4.02QTNYY205 pKa = 9.87VPNPKK210 pKa = 10.2GVISLAIEE218 pKa = 4.16DD219 pKa = 4.84LKK221 pKa = 11.23PLINFDD227 pKa = 3.83PYY229 pKa = 11.62QMVRR233 pKa = 11.84LLGLSPMIEE242 pKa = 4.4DD243 pKa = 6.03PIPAPSKK250 pKa = 9.58TRR252 pKa = 11.84SILPAKK258 pKa = 9.99FFKK261 pKa = 10.75KK262 pKa = 10.49
Molecular weight: 30.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4163 |
262 |
2328 |
832.6 |
86.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.02 ± 1.042 | 1.417 ± 0.21 |
4.228 ± 0.32 | 4.66 ± 0.271 |
3.579 ± 0.393 | 4.588 ± 0.414 |
2.618 ± 0.137 | 6.101 ± 0.378 |
4.3 ± 0.751 | 10.545 ± 0.427 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.402 ± 0.321 | 4.396 ± 0.261 |
5.213 ± 0.498 | 3.843 ± 0.364 |
5.237 ± 0.064 | 7.278 ± 0.848 |
5.525 ± 0.599 | 4.66 ± 0.578 |
1.513 ± 0.254 | 3.603 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
