
Lake Sarah-associated circular virus-3
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
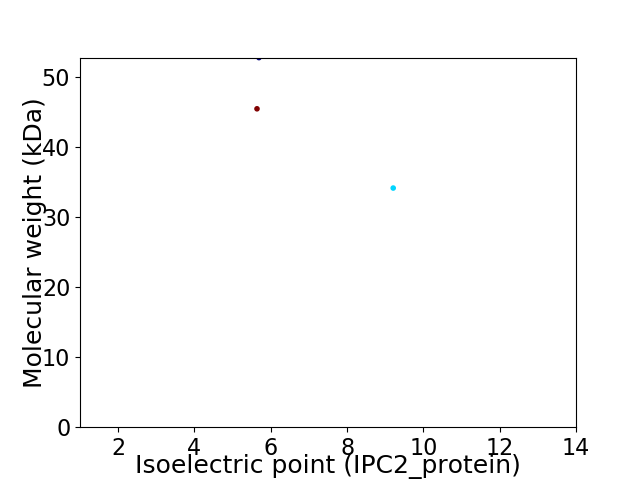
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQJ5|A0A140AQJ5_9VIRU Uncharacterized protein OS=Lake Sarah-associated circular virus-3 OX=1685757 PE=4 SV=1
MM1 pKa = 7.47NLLLCLCNVTQFNSMSSDD19 pKa = 3.29TVKK22 pKa = 10.92CGVATLDD29 pKa = 3.7PFRR32 pKa = 11.84PAAKK36 pKa = 9.79GACLPTGSCPATYY49 pKa = 9.81RR50 pKa = 11.84ASAVIRR56 pKa = 11.84VDD58 pKa = 4.0GVLGPQGQGIIMVNPSVASDD78 pKa = 4.41AACLWYY84 pKa = 11.1SNGSTFTQSPVVIGNSAYY102 pKa = 11.11VNGTGLTGPVNGLPTLITPGLTQATATSLPFQSDD136 pKa = 2.84AMFGFNASTALPLPASGPPQVRR158 pKa = 11.84ARR160 pKa = 11.84IVSCGVKK167 pKa = 8.76ITFSGTTLNDD177 pKa = 3.2GGVAYY182 pKa = 10.48CLVDD186 pKa = 3.78PQHH189 pKa = 6.65EE190 pKa = 4.21NLIEE194 pKa = 4.11LGINNYY200 pKa = 10.19LSLFTSCKK208 pKa = 8.77IQRR211 pKa = 11.84LSLRR215 pKa = 11.84DD216 pKa = 3.82TIALNLAPVTRR227 pKa = 11.84NQQDD231 pKa = 3.2LSSAYY236 pKa = 10.49DD237 pKa = 3.73EE238 pKa = 4.9VPLGSIFGGSLNGGYY253 pKa = 9.64IYY255 pKa = 10.92SPFSMPNPTSNFGYY269 pKa = 10.18GYY271 pKa = 9.43PNVGQSTQSCVPCDD285 pKa = 3.51AFNCAKK291 pKa = 10.42AASTLLYY298 pKa = 9.72PLSRR302 pKa = 11.84RR303 pKa = 11.84NQRR306 pKa = 11.84VIPWTVNNSTTTGLWSGSVAGGGLITWTSIAPDD339 pKa = 3.47VGASGMIMSGTVSGLGTIDD358 pKa = 3.38VEE360 pKa = 4.4GVIWYY365 pKa = 9.53RR366 pKa = 11.84YY367 pKa = 10.01DD368 pKa = 3.29NNSWYY373 pKa = 10.43FSLTSGVPTITPFNFTNWAITGYY396 pKa = 8.6WCEE399 pKa = 3.86PSVIGAVIIQAGTAMAGQTFHH420 pKa = 7.84IEE422 pKa = 4.15YY423 pKa = 10.05VVHH426 pKa = 6.73CEE428 pKa = 3.77YY429 pKa = 11.14SGVGVQGRR437 pKa = 11.84TEE439 pKa = 4.27NIIPDD444 pKa = 3.97PEE446 pKa = 4.89GLAGVHH452 pKa = 6.51AVLDD456 pKa = 4.04LCRR459 pKa = 11.84EE460 pKa = 4.06SAGQHH465 pKa = 4.2EE466 pKa = 4.68RR467 pKa = 11.84ANIKK471 pKa = 10.69DD472 pKa = 3.87FVSAASSKK480 pKa = 10.16LVKK483 pKa = 10.49SGALAVKK490 pKa = 9.75LGEE493 pKa = 4.24VASVLGKK500 pKa = 10.46RR501 pKa = 11.84FNN503 pKa = 3.58
MM1 pKa = 7.47NLLLCLCNVTQFNSMSSDD19 pKa = 3.29TVKK22 pKa = 10.92CGVATLDD29 pKa = 3.7PFRR32 pKa = 11.84PAAKK36 pKa = 9.79GACLPTGSCPATYY49 pKa = 9.81RR50 pKa = 11.84ASAVIRR56 pKa = 11.84VDD58 pKa = 4.0GVLGPQGQGIIMVNPSVASDD78 pKa = 4.41AACLWYY84 pKa = 11.1SNGSTFTQSPVVIGNSAYY102 pKa = 11.11VNGTGLTGPVNGLPTLITPGLTQATATSLPFQSDD136 pKa = 2.84AMFGFNASTALPLPASGPPQVRR158 pKa = 11.84ARR160 pKa = 11.84IVSCGVKK167 pKa = 8.76ITFSGTTLNDD177 pKa = 3.2GGVAYY182 pKa = 10.48CLVDD186 pKa = 3.78PQHH189 pKa = 6.65EE190 pKa = 4.21NLIEE194 pKa = 4.11LGINNYY200 pKa = 10.19LSLFTSCKK208 pKa = 8.77IQRR211 pKa = 11.84LSLRR215 pKa = 11.84DD216 pKa = 3.82TIALNLAPVTRR227 pKa = 11.84NQQDD231 pKa = 3.2LSSAYY236 pKa = 10.49DD237 pKa = 3.73EE238 pKa = 4.9VPLGSIFGGSLNGGYY253 pKa = 9.64IYY255 pKa = 10.92SPFSMPNPTSNFGYY269 pKa = 10.18GYY271 pKa = 9.43PNVGQSTQSCVPCDD285 pKa = 3.51AFNCAKK291 pKa = 10.42AASTLLYY298 pKa = 9.72PLSRR302 pKa = 11.84RR303 pKa = 11.84NQRR306 pKa = 11.84VIPWTVNNSTTTGLWSGSVAGGGLITWTSIAPDD339 pKa = 3.47VGASGMIMSGTVSGLGTIDD358 pKa = 3.38VEE360 pKa = 4.4GVIWYY365 pKa = 9.53RR366 pKa = 11.84YY367 pKa = 10.01DD368 pKa = 3.29NNSWYY373 pKa = 10.43FSLTSGVPTITPFNFTNWAITGYY396 pKa = 8.6WCEE399 pKa = 3.86PSVIGAVIIQAGTAMAGQTFHH420 pKa = 7.84IEE422 pKa = 4.15YY423 pKa = 10.05VVHH426 pKa = 6.73CEE428 pKa = 3.77YY429 pKa = 11.14SGVGVQGRR437 pKa = 11.84TEE439 pKa = 4.27NIIPDD444 pKa = 3.97PEE446 pKa = 4.89GLAGVHH452 pKa = 6.51AVLDD456 pKa = 4.04LCRR459 pKa = 11.84EE460 pKa = 4.06SAGQHH465 pKa = 4.2EE466 pKa = 4.68RR467 pKa = 11.84ANIKK471 pKa = 10.69DD472 pKa = 3.87FVSAASSKK480 pKa = 10.16LVKK483 pKa = 10.49SGALAVKK490 pKa = 9.75LGEE493 pKa = 4.24VASVLGKK500 pKa = 10.46RR501 pKa = 11.84FNN503 pKa = 3.58
Molecular weight: 52.77 kDa
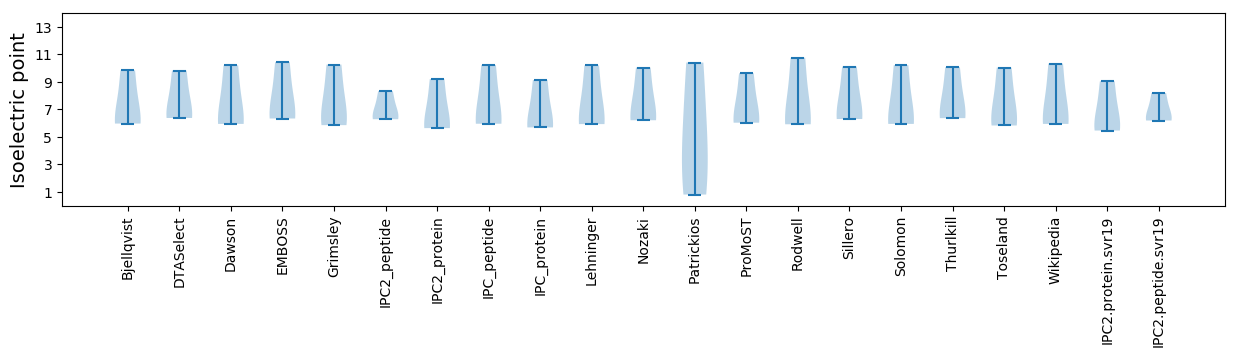
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9I2|A0A126G9I2_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-3 OX=1685757 PE=3 SV=1
MM1 pKa = 7.67AKK3 pKa = 10.08YY4 pKa = 9.26FKK6 pKa = 9.87PSKK9 pKa = 10.2KK10 pKa = 9.93FPTRR14 pKa = 11.84EE15 pKa = 3.3HH16 pKa = 5.56YY17 pKa = 10.39RR18 pKa = 11.84NYY20 pKa = 10.11RR21 pKa = 11.84EE22 pKa = 3.92AMGIRR27 pKa = 11.84GKK29 pKa = 10.58KK30 pKa = 8.23SSVYY34 pKa = 10.21KK35 pKa = 10.46RR36 pKa = 11.84LRR38 pKa = 11.84GNKK41 pKa = 7.58LTRR44 pKa = 11.84STRR47 pKa = 11.84YY48 pKa = 8.59KK49 pKa = 9.81QKK51 pKa = 10.47KK52 pKa = 9.25FGFKK56 pKa = 9.74RR57 pKa = 11.84SSYY60 pKa = 11.37AKK62 pKa = 10.15GLKK65 pKa = 10.25LPLSTTCPKK74 pKa = 11.1NMLFLSNAFIEE85 pKa = 4.35YY86 pKa = 10.06FKK88 pKa = 10.93ISLKK92 pKa = 9.9MLNDD96 pKa = 3.57EE97 pKa = 4.69PFKK100 pKa = 11.62DD101 pKa = 3.57MLALVSRR108 pKa = 11.84IPTYY112 pKa = 10.36RR113 pKa = 11.84ALDD116 pKa = 3.62AFATLITCTPFSKK129 pKa = 9.74YY130 pKa = 7.97TAVGDD135 pKa = 4.25AVQEE139 pKa = 4.04MCRR142 pKa = 11.84QFLVLFKK149 pKa = 10.22TAIAHH154 pKa = 6.53AEE156 pKa = 4.12SEE158 pKa = 4.3LLVPGAHH165 pKa = 4.55QKK167 pKa = 8.88MRR169 pKa = 11.84EE170 pKa = 4.01VEE172 pKa = 3.95HH173 pKa = 6.98ALRR176 pKa = 11.84EE177 pKa = 4.31HH178 pKa = 5.69TKK180 pKa = 9.74EE181 pKa = 4.09ANVEE185 pKa = 3.95LAEE188 pKa = 5.1HH189 pKa = 6.71ILHH192 pKa = 6.95EE193 pKa = 4.67RR194 pKa = 11.84MQQANEE200 pKa = 3.76QMEE203 pKa = 4.55KK204 pKa = 10.45FGAHH208 pKa = 6.25MGDD211 pKa = 3.1KK212 pKa = 10.27VGKK215 pKa = 9.31LARR218 pKa = 11.84ARR220 pKa = 11.84NEE222 pKa = 3.53VMEE225 pKa = 4.21RR226 pKa = 11.84AKK228 pKa = 11.09VEE230 pKa = 4.28MVDD233 pKa = 3.84GTQSAAAQSGDD244 pKa = 3.59MYY246 pKa = 10.73HH247 pKa = 7.0KK248 pKa = 9.89SHH250 pKa = 6.55HH251 pKa = 6.69HH252 pKa = 5.29GHH254 pKa = 6.49HH255 pKa = 6.37KK256 pKa = 10.53GINLKK261 pKa = 10.37SVSRR265 pKa = 11.84ALNHH269 pKa = 6.51AEE271 pKa = 3.85KK272 pKa = 10.6SVMSGAEE279 pKa = 3.85SVMRR283 pKa = 11.84TVTQTVASDD292 pKa = 3.9PEE294 pKa = 4.42SVAMFMM300 pKa = 5.4
MM1 pKa = 7.67AKK3 pKa = 10.08YY4 pKa = 9.26FKK6 pKa = 9.87PSKK9 pKa = 10.2KK10 pKa = 9.93FPTRR14 pKa = 11.84EE15 pKa = 3.3HH16 pKa = 5.56YY17 pKa = 10.39RR18 pKa = 11.84NYY20 pKa = 10.11RR21 pKa = 11.84EE22 pKa = 3.92AMGIRR27 pKa = 11.84GKK29 pKa = 10.58KK30 pKa = 8.23SSVYY34 pKa = 10.21KK35 pKa = 10.46RR36 pKa = 11.84LRR38 pKa = 11.84GNKK41 pKa = 7.58LTRR44 pKa = 11.84STRR47 pKa = 11.84YY48 pKa = 8.59KK49 pKa = 9.81QKK51 pKa = 10.47KK52 pKa = 9.25FGFKK56 pKa = 9.74RR57 pKa = 11.84SSYY60 pKa = 11.37AKK62 pKa = 10.15GLKK65 pKa = 10.25LPLSTTCPKK74 pKa = 11.1NMLFLSNAFIEE85 pKa = 4.35YY86 pKa = 10.06FKK88 pKa = 10.93ISLKK92 pKa = 9.9MLNDD96 pKa = 3.57EE97 pKa = 4.69PFKK100 pKa = 11.62DD101 pKa = 3.57MLALVSRR108 pKa = 11.84IPTYY112 pKa = 10.36RR113 pKa = 11.84ALDD116 pKa = 3.62AFATLITCTPFSKK129 pKa = 9.74YY130 pKa = 7.97TAVGDD135 pKa = 4.25AVQEE139 pKa = 4.04MCRR142 pKa = 11.84QFLVLFKK149 pKa = 10.22TAIAHH154 pKa = 6.53AEE156 pKa = 4.12SEE158 pKa = 4.3LLVPGAHH165 pKa = 4.55QKK167 pKa = 8.88MRR169 pKa = 11.84EE170 pKa = 4.01VEE172 pKa = 3.95HH173 pKa = 6.98ALRR176 pKa = 11.84EE177 pKa = 4.31HH178 pKa = 5.69TKK180 pKa = 9.74EE181 pKa = 4.09ANVEE185 pKa = 3.95LAEE188 pKa = 5.1HH189 pKa = 6.71ILHH192 pKa = 6.95EE193 pKa = 4.67RR194 pKa = 11.84MQQANEE200 pKa = 3.76QMEE203 pKa = 4.55KK204 pKa = 10.45FGAHH208 pKa = 6.25MGDD211 pKa = 3.1KK212 pKa = 10.27VGKK215 pKa = 9.31LARR218 pKa = 11.84ARR220 pKa = 11.84NEE222 pKa = 3.53VMEE225 pKa = 4.21RR226 pKa = 11.84AKK228 pKa = 11.09VEE230 pKa = 4.28MVDD233 pKa = 3.84GTQSAAAQSGDD244 pKa = 3.59MYY246 pKa = 10.73HH247 pKa = 7.0KK248 pKa = 9.89SHH250 pKa = 6.55HH251 pKa = 6.69HH252 pKa = 5.29GHH254 pKa = 6.49HH255 pKa = 6.37KK256 pKa = 10.53GINLKK261 pKa = 10.37SVSRR265 pKa = 11.84ALNHH269 pKa = 6.51AEE271 pKa = 3.85KK272 pKa = 10.6SVMSGAEE279 pKa = 3.85SVMRR283 pKa = 11.84TVTQTVASDD292 pKa = 3.9PEE294 pKa = 4.42SVAMFMM300 pKa = 5.4
Molecular weight: 34.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1202 |
300 |
503 |
400.7 |
44.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.987 ± 0.898 | 2.163 ± 0.445 |
4.576 ± 1.21 | 4.576 ± 1.169 |
5.075 ± 1.022 | 7.404 ± 1.801 |
3.661 ± 1.492 | 5.408 ± 0.841 |
4.576 ± 1.627 | 7.82 ± 0.31 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.329 ± 0.919 | 4.077 ± 1.079 |
5.74 ± 0.779 | 3.078 ± 0.367 |
4.992 ± 0.962 | 7.737 ± 1.199 |
7.571 ± 0.63 | 7.072 ± 0.683 |
1.082 ± 0.323 | 3.078 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
