
Adana virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Adana phlebovirus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
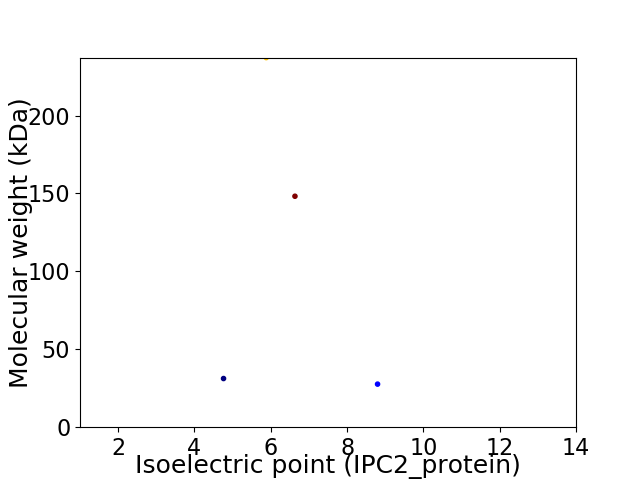
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
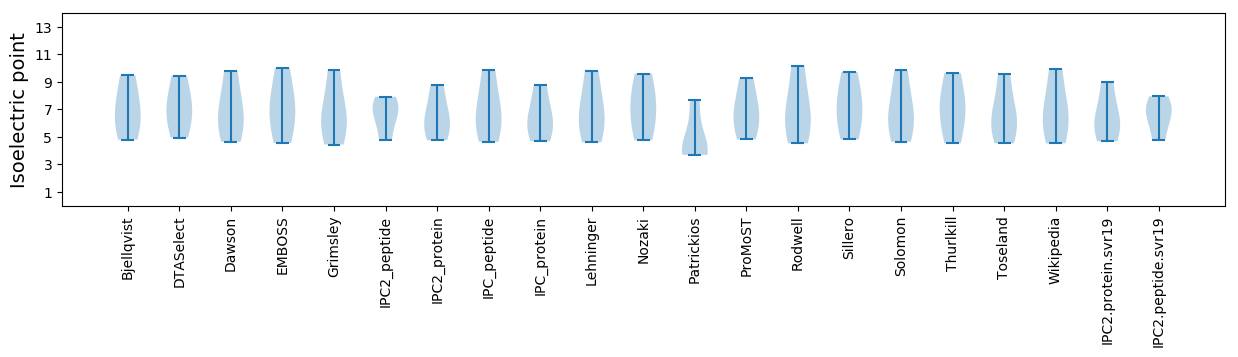
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5B2U6|A0A0C5B2U6_9VIRU Nonstructural protein OS=Adana virus OX=1611877 PE=4 SV=1
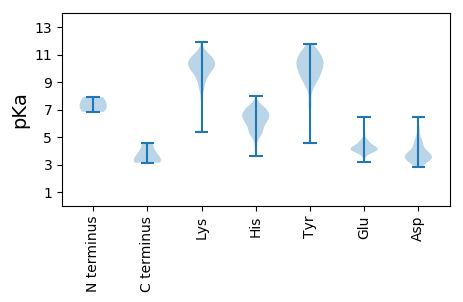
MM1 pKa = 7.13SLKK4 pKa = 10.18YY5 pKa = 10.51LCDD8 pKa = 5.33RR9 pKa = 11.84ITPTYY14 pKa = 9.28DD15 pKa = 3.14YY16 pKa = 10.35PVIGEE21 pKa = 4.2VVVSFEE27 pKa = 4.29AVNSQVDD34 pKa = 4.35LPVCVYY40 pKa = 11.34DD41 pKa = 4.71DD42 pKa = 5.83LEE44 pKa = 5.32FEE46 pKa = 4.36LAGFRR51 pKa = 11.84LSLAYY56 pKa = 9.85KK57 pKa = 7.77NTLYY61 pKa = 10.93EE62 pKa = 4.9FIDD65 pKa = 3.64NDD67 pKa = 3.96EE68 pKa = 4.55IPWRR72 pKa = 11.84WGPHH76 pKa = 3.62MWEE79 pKa = 4.26SRR81 pKa = 11.84VTMDD85 pKa = 5.58EE86 pKa = 4.03IPLLDD91 pKa = 3.74SLMKK95 pKa = 9.94TFQGLPVTDD104 pKa = 4.51LMDD107 pKa = 4.55PRR109 pKa = 11.84LHH111 pKa = 5.96FTRR114 pKa = 11.84NALSWPIGRR123 pKa = 11.84PSLKK127 pKa = 10.13AFRR130 pKa = 11.84ILYY133 pKa = 7.11PCRR136 pKa = 11.84FVGFSCSKK144 pKa = 10.72RR145 pKa = 11.84EE146 pKa = 3.6LAEE149 pKa = 4.21YY150 pKa = 10.15IFEE153 pKa = 4.2VTKK156 pKa = 10.76AVNLEE161 pKa = 4.06CGIVQMAKK169 pKa = 9.93MIRR172 pKa = 11.84STAQSMGVQEE182 pKa = 4.62SLVPCKK188 pKa = 11.02DD189 pKa = 3.26MVKK192 pKa = 10.06CICILQFIKK201 pKa = 10.06MIKK204 pKa = 10.07GFQLDD209 pKa = 3.32KK210 pKa = 10.61TRR212 pKa = 11.84NVLFGGALIPFLHH225 pKa = 6.95ACTEE229 pKa = 3.85ALTRR233 pKa = 11.84DD234 pKa = 4.05FPGLWRR240 pKa = 11.84AWNPQADD247 pKa = 3.82IMDD250 pKa = 6.47LITADD255 pKa = 4.65PYY257 pKa = 11.76DD258 pKa = 4.11SGISTDD264 pKa = 3.92SEE266 pKa = 3.96FDD268 pKa = 3.15SDD270 pKa = 4.69IEE272 pKa = 4.3VV273 pKa = 3.14
MM1 pKa = 7.13SLKK4 pKa = 10.18YY5 pKa = 10.51LCDD8 pKa = 5.33RR9 pKa = 11.84ITPTYY14 pKa = 9.28DD15 pKa = 3.14YY16 pKa = 10.35PVIGEE21 pKa = 4.2VVVSFEE27 pKa = 4.29AVNSQVDD34 pKa = 4.35LPVCVYY40 pKa = 11.34DD41 pKa = 4.71DD42 pKa = 5.83LEE44 pKa = 5.32FEE46 pKa = 4.36LAGFRR51 pKa = 11.84LSLAYY56 pKa = 9.85KK57 pKa = 7.77NTLYY61 pKa = 10.93EE62 pKa = 4.9FIDD65 pKa = 3.64NDD67 pKa = 3.96EE68 pKa = 4.55IPWRR72 pKa = 11.84WGPHH76 pKa = 3.62MWEE79 pKa = 4.26SRR81 pKa = 11.84VTMDD85 pKa = 5.58EE86 pKa = 4.03IPLLDD91 pKa = 3.74SLMKK95 pKa = 9.94TFQGLPVTDD104 pKa = 4.51LMDD107 pKa = 4.55PRR109 pKa = 11.84LHH111 pKa = 5.96FTRR114 pKa = 11.84NALSWPIGRR123 pKa = 11.84PSLKK127 pKa = 10.13AFRR130 pKa = 11.84ILYY133 pKa = 7.11PCRR136 pKa = 11.84FVGFSCSKK144 pKa = 10.72RR145 pKa = 11.84EE146 pKa = 3.6LAEE149 pKa = 4.21YY150 pKa = 10.15IFEE153 pKa = 4.2VTKK156 pKa = 10.76AVNLEE161 pKa = 4.06CGIVQMAKK169 pKa = 9.93MIRR172 pKa = 11.84STAQSMGVQEE182 pKa = 4.62SLVPCKK188 pKa = 11.02DD189 pKa = 3.26MVKK192 pKa = 10.06CICILQFIKK201 pKa = 10.06MIKK204 pKa = 10.07GFQLDD209 pKa = 3.32KK210 pKa = 10.61TRR212 pKa = 11.84NVLFGGALIPFLHH225 pKa = 6.95ACTEE229 pKa = 3.85ALTRR233 pKa = 11.84DD234 pKa = 4.05FPGLWRR240 pKa = 11.84AWNPQADD247 pKa = 3.82IMDD250 pKa = 6.47LITADD255 pKa = 4.65PYY257 pKa = 11.76DD258 pKa = 4.11SGISTDD264 pKa = 3.92SEE266 pKa = 3.96FDD268 pKa = 3.15SDD270 pKa = 4.69IEE272 pKa = 4.3VV273 pKa = 3.14
Molecular weight: 31.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5ARY4|A0A0C5ARY4_9VIRU Glycoprotein complex OS=Adana virus OX=1611877 PE=4 SV=1
MM1 pKa = 7.9SSQEE5 pKa = 3.82DD6 pKa = 3.8FARR9 pKa = 11.84IAVEE13 pKa = 4.14FANEE17 pKa = 3.84AVDD20 pKa = 3.66TNQILEE26 pKa = 4.31LVRR29 pKa = 11.84EE30 pKa = 4.56FAYY33 pKa = 10.2QGYY36 pKa = 9.31DD37 pKa = 2.91AARR40 pKa = 11.84VIEE43 pKa = 4.02LVRR46 pKa = 11.84SKK48 pKa = 11.18GGEE51 pKa = 3.52NWKK54 pKa = 10.51NDD56 pKa = 3.3VKK58 pKa = 10.21TLIVIALTRR67 pKa = 11.84GNKK70 pKa = 8.56PSKK73 pKa = 9.73ILSKK77 pKa = 8.12MTPDD81 pKa = 3.18AGRR84 pKa = 11.84KK85 pKa = 8.35FRR87 pKa = 11.84EE88 pKa = 3.59LVMRR92 pKa = 11.84YY93 pKa = 8.63GLKK96 pKa = 10.28SGNPGRR102 pKa = 11.84DD103 pKa = 3.46DD104 pKa = 3.39LTLARR109 pKa = 11.84IASAFAAWTVQAIKK123 pKa = 10.63VVEE126 pKa = 4.26QYY128 pKa = 11.52LPVTGTAMDD137 pKa = 4.91DD138 pKa = 3.85LSPGYY143 pKa = 8.61PRR145 pKa = 11.84PMMHH149 pKa = 7.08PCFAGLIDD157 pKa = 3.65NTLPEE162 pKa = 4.03EE163 pKa = 4.31TYY165 pKa = 10.49RR166 pKa = 11.84AILRR170 pKa = 11.84AHH172 pKa = 6.52SLFLDD177 pKa = 3.48QFSRR181 pKa = 11.84TINPQMRR188 pKa = 11.84GKK190 pKa = 9.67PKK192 pKa = 10.72SEE194 pKa = 3.83VAKK197 pKa = 10.87SFEE200 pKa = 4.22QPLQAAVNSRR210 pKa = 11.84FLSSEE215 pKa = 4.07SKK217 pKa = 10.59RR218 pKa = 11.84KK219 pKa = 9.35ILRR222 pKa = 11.84STGIIDD228 pKa = 4.6SNLKK232 pKa = 8.9PAPAVEE238 pKa = 4.12HH239 pKa = 6.1AAKK242 pKa = 10.41KK243 pKa = 9.26FLEE246 pKa = 4.42MAA248 pKa = 4.6
MM1 pKa = 7.9SSQEE5 pKa = 3.82DD6 pKa = 3.8FARR9 pKa = 11.84IAVEE13 pKa = 4.14FANEE17 pKa = 3.84AVDD20 pKa = 3.66TNQILEE26 pKa = 4.31LVRR29 pKa = 11.84EE30 pKa = 4.56FAYY33 pKa = 10.2QGYY36 pKa = 9.31DD37 pKa = 2.91AARR40 pKa = 11.84VIEE43 pKa = 4.02LVRR46 pKa = 11.84SKK48 pKa = 11.18GGEE51 pKa = 3.52NWKK54 pKa = 10.51NDD56 pKa = 3.3VKK58 pKa = 10.21TLIVIALTRR67 pKa = 11.84GNKK70 pKa = 8.56PSKK73 pKa = 9.73ILSKK77 pKa = 8.12MTPDD81 pKa = 3.18AGRR84 pKa = 11.84KK85 pKa = 8.35FRR87 pKa = 11.84EE88 pKa = 3.59LVMRR92 pKa = 11.84YY93 pKa = 8.63GLKK96 pKa = 10.28SGNPGRR102 pKa = 11.84DD103 pKa = 3.46DD104 pKa = 3.39LTLARR109 pKa = 11.84IASAFAAWTVQAIKK123 pKa = 10.63VVEE126 pKa = 4.26QYY128 pKa = 11.52LPVTGTAMDD137 pKa = 4.91DD138 pKa = 3.85LSPGYY143 pKa = 8.61PRR145 pKa = 11.84PMMHH149 pKa = 7.08PCFAGLIDD157 pKa = 3.65NTLPEE162 pKa = 4.03EE163 pKa = 4.31TYY165 pKa = 10.49RR166 pKa = 11.84AILRR170 pKa = 11.84AHH172 pKa = 6.52SLFLDD177 pKa = 3.48QFSRR181 pKa = 11.84TINPQMRR188 pKa = 11.84GKK190 pKa = 9.67PKK192 pKa = 10.72SEE194 pKa = 3.83VAKK197 pKa = 10.87SFEE200 pKa = 4.22QPLQAAVNSRR210 pKa = 11.84FLSSEE215 pKa = 4.07SKK217 pKa = 10.59RR218 pKa = 11.84KK219 pKa = 9.35ILRR222 pKa = 11.84STGIIDD228 pKa = 4.6SNLKK232 pKa = 8.9PAPAVEE238 pKa = 4.12HH239 pKa = 6.1AAKK242 pKa = 10.41KK243 pKa = 9.26FLEE246 pKa = 4.42MAA248 pKa = 4.6
Molecular weight: 27.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3952 |
248 |
2096 |
988.0 |
111.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.452 ± 0.485 | 2.657 ± 0.772 |
5.541 ± 0.454 | 6.705 ± 0.349 |
4.352 ± 0.357 | 5.719 ± 0.201 |
2.429 ± 0.312 | 6.402 ± 0.148 |
5.896 ± 0.344 | 8.856 ± 0.375 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.214 ± 0.4 | 3.669 ± 0.363 |
4.352 ± 0.329 | 3.087 ± 0.092 |
5.693 ± 0.212 | 8.401 ± 0.433 |
5.845 ± 0.36 | 6.68 ± 0.359 |
1.265 ± 0.173 | 2.783 ± 0.094 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
