
Rhizobium sp. CF080
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
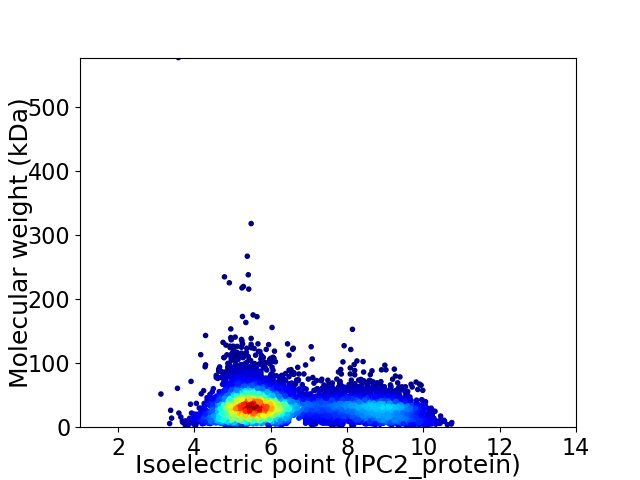
Virtual 2D-PAGE plot for 6670 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W6W9F4|W6W9F4_9RHIZ Outer membrane protein assembly factor BamA OS=Rhizobium sp. CF080 OX=1144310 GN=bamA PE=3 SV=1
MM1 pKa = 7.38ATNSDD6 pKa = 3.85NFDD9 pKa = 3.43LTSWKK14 pKa = 10.38LCLPIDD20 pKa = 4.21EE21 pKa = 5.54DD22 pKa = 3.95GGTIGTALEE31 pKa = 4.08ILDD34 pKa = 4.03LSGFEE39 pKa = 4.12HH40 pKa = 7.22SSYY43 pKa = 10.8FYY45 pKa = 10.37TADD48 pKa = 3.55DD49 pKa = 3.69GAMVFRR55 pKa = 11.84AITEE59 pKa = 4.08GALTKK64 pKa = 10.05GTTCARR70 pKa = 11.84SEE72 pKa = 3.82LRR74 pKa = 11.84EE75 pKa = 4.11MNGHH79 pKa = 6.33SLASWTLDD87 pKa = 2.92EE88 pKa = 5.27GGTMTATLKK97 pKa = 10.2IDD99 pKa = 4.02EE100 pKa = 5.25APHH103 pKa = 4.97TTNGAEE109 pKa = 3.88GRR111 pKa = 11.84IVVGQIHH118 pKa = 6.48GSEE121 pKa = 4.34DD122 pKa = 3.51EE123 pKa = 4.16LVRR126 pKa = 11.84LYY128 pKa = 10.4WEE130 pKa = 4.65DD131 pKa = 3.85GEE133 pKa = 5.87VYY135 pKa = 10.49FKK137 pKa = 10.99SDD139 pKa = 3.71KK140 pKa = 11.53GGDD143 pKa = 3.8GNDD146 pKa = 3.38SLKK149 pKa = 10.75FALTNAGGDD158 pKa = 3.66TPQISLGDD166 pKa = 3.59QFSYY170 pKa = 10.66KK171 pKa = 9.55IDD173 pKa = 3.44VHH175 pKa = 7.81GDD177 pKa = 3.12TLTVIVYY184 pKa = 10.71ADD186 pKa = 3.54GDD188 pKa = 4.3TYY190 pKa = 11.49TSVTTVSSAWDD201 pKa = 3.55GEE203 pKa = 4.22QFYY206 pKa = 10.43FKK208 pKa = 10.86AGAYY212 pKa = 9.82LGNNEE217 pKa = 4.44TNSTGAGQVSFYY229 pKa = 11.18GFDD232 pKa = 3.78FSHH235 pKa = 6.07EE236 pKa = 4.28TGEE239 pKa = 4.29GLDD242 pKa = 4.02GLVSTATANDD252 pKa = 3.69DD253 pKa = 3.51SSTYY257 pKa = 10.95SVDD260 pKa = 3.26STGTIGNDD268 pKa = 2.84VLTGGTLADD277 pKa = 3.66VIYY280 pKa = 10.44SYY282 pKa = 11.4GGNDD286 pKa = 3.31VVRR289 pKa = 11.84AGAGDD294 pKa = 3.61DD295 pKa = 3.45RR296 pKa = 11.84VVGGSGADD304 pKa = 3.22KK305 pKa = 11.18LLGQDD310 pKa = 3.45GADD313 pKa = 3.75TIFAGADD320 pKa = 3.32DD321 pKa = 4.26DD322 pKa = 4.61VVYY325 pKa = 10.84GGDD328 pKa = 3.85GDD330 pKa = 4.0DD331 pKa = 4.79TIDD334 pKa = 3.96GGAGNDD340 pKa = 3.82TLKK343 pKa = 11.41GEE345 pKa = 4.42AGANRR350 pKa = 11.84ITGGEE355 pKa = 4.14GDD357 pKa = 3.64DD358 pKa = 4.04TIYY361 pKa = 11.24GGVGADD367 pKa = 4.63DD368 pKa = 4.51LSGDD372 pKa = 3.74AGEE375 pKa = 4.38DD376 pKa = 3.52TIVAGEE382 pKa = 4.3GDD384 pKa = 4.83DD385 pKa = 3.91ILSGGADD392 pKa = 3.37NDD394 pKa = 3.61KK395 pKa = 11.22LYY397 pKa = 11.07GGEE400 pKa = 4.2GADD403 pKa = 3.33RR404 pKa = 11.84LYY406 pKa = 11.28GQGGDD411 pKa = 4.36DD412 pKa = 3.61VLIGQGGADD421 pKa = 3.75KK422 pKa = 11.16LVGADD427 pKa = 4.87GNDD430 pKa = 3.35TLYY433 pKa = 11.03GYY435 pKa = 10.91SDD437 pKa = 3.51NDD439 pKa = 3.55KK440 pKa = 10.99LYY442 pKa = 11.38GDD444 pKa = 4.5AGSDD448 pKa = 3.4KK449 pKa = 11.01LVGGDD454 pKa = 4.92GDD456 pKa = 3.77DD457 pKa = 3.57TLYY460 pKa = 11.36GGAGDD465 pKa = 4.13DD466 pKa = 4.17RR467 pKa = 11.84LYY469 pKa = 11.32ADD471 pKa = 4.43EE472 pKa = 5.4GADD475 pKa = 3.42NLYY478 pKa = 10.99GGTGADD484 pKa = 2.91TFVFTWLDD492 pKa = 3.56PSTLTSGGRR501 pKa = 11.84DD502 pKa = 3.57DD503 pKa = 5.18IYY505 pKa = 11.24QFSEE509 pKa = 4.02ADD511 pKa = 3.1GDD513 pKa = 4.2IINLSGIDD521 pKa = 3.91ANPTVNGGQAFDD533 pKa = 4.92FIGTNAFSEE542 pKa = 4.58QVSEE546 pKa = 3.92LRR548 pKa = 11.84YY549 pKa = 10.24VNTGSEE555 pKa = 4.26TYY557 pKa = 10.01IYY559 pKa = 11.06GDD561 pKa = 3.39INGDD565 pKa = 3.31GAADD569 pKa = 4.06FSIHH573 pKa = 6.01VDD575 pKa = 3.61GVVEE579 pKa = 4.06LQSIDD584 pKa = 4.05FILL587 pKa = 4.79
MM1 pKa = 7.38ATNSDD6 pKa = 3.85NFDD9 pKa = 3.43LTSWKK14 pKa = 10.38LCLPIDD20 pKa = 4.21EE21 pKa = 5.54DD22 pKa = 3.95GGTIGTALEE31 pKa = 4.08ILDD34 pKa = 4.03LSGFEE39 pKa = 4.12HH40 pKa = 7.22SSYY43 pKa = 10.8FYY45 pKa = 10.37TADD48 pKa = 3.55DD49 pKa = 3.69GAMVFRR55 pKa = 11.84AITEE59 pKa = 4.08GALTKK64 pKa = 10.05GTTCARR70 pKa = 11.84SEE72 pKa = 3.82LRR74 pKa = 11.84EE75 pKa = 4.11MNGHH79 pKa = 6.33SLASWTLDD87 pKa = 2.92EE88 pKa = 5.27GGTMTATLKK97 pKa = 10.2IDD99 pKa = 4.02EE100 pKa = 5.25APHH103 pKa = 4.97TTNGAEE109 pKa = 3.88GRR111 pKa = 11.84IVVGQIHH118 pKa = 6.48GSEE121 pKa = 4.34DD122 pKa = 3.51EE123 pKa = 4.16LVRR126 pKa = 11.84LYY128 pKa = 10.4WEE130 pKa = 4.65DD131 pKa = 3.85GEE133 pKa = 5.87VYY135 pKa = 10.49FKK137 pKa = 10.99SDD139 pKa = 3.71KK140 pKa = 11.53GGDD143 pKa = 3.8GNDD146 pKa = 3.38SLKK149 pKa = 10.75FALTNAGGDD158 pKa = 3.66TPQISLGDD166 pKa = 3.59QFSYY170 pKa = 10.66KK171 pKa = 9.55IDD173 pKa = 3.44VHH175 pKa = 7.81GDD177 pKa = 3.12TLTVIVYY184 pKa = 10.71ADD186 pKa = 3.54GDD188 pKa = 4.3TYY190 pKa = 11.49TSVTTVSSAWDD201 pKa = 3.55GEE203 pKa = 4.22QFYY206 pKa = 10.43FKK208 pKa = 10.86AGAYY212 pKa = 9.82LGNNEE217 pKa = 4.44TNSTGAGQVSFYY229 pKa = 11.18GFDD232 pKa = 3.78FSHH235 pKa = 6.07EE236 pKa = 4.28TGEE239 pKa = 4.29GLDD242 pKa = 4.02GLVSTATANDD252 pKa = 3.69DD253 pKa = 3.51SSTYY257 pKa = 10.95SVDD260 pKa = 3.26STGTIGNDD268 pKa = 2.84VLTGGTLADD277 pKa = 3.66VIYY280 pKa = 10.44SYY282 pKa = 11.4GGNDD286 pKa = 3.31VVRR289 pKa = 11.84AGAGDD294 pKa = 3.61DD295 pKa = 3.45RR296 pKa = 11.84VVGGSGADD304 pKa = 3.22KK305 pKa = 11.18LLGQDD310 pKa = 3.45GADD313 pKa = 3.75TIFAGADD320 pKa = 3.32DD321 pKa = 4.26DD322 pKa = 4.61VVYY325 pKa = 10.84GGDD328 pKa = 3.85GDD330 pKa = 4.0DD331 pKa = 4.79TIDD334 pKa = 3.96GGAGNDD340 pKa = 3.82TLKK343 pKa = 11.41GEE345 pKa = 4.42AGANRR350 pKa = 11.84ITGGEE355 pKa = 4.14GDD357 pKa = 3.64DD358 pKa = 4.04TIYY361 pKa = 11.24GGVGADD367 pKa = 4.63DD368 pKa = 4.51LSGDD372 pKa = 3.74AGEE375 pKa = 4.38DD376 pKa = 3.52TIVAGEE382 pKa = 4.3GDD384 pKa = 4.83DD385 pKa = 3.91ILSGGADD392 pKa = 3.37NDD394 pKa = 3.61KK395 pKa = 11.22LYY397 pKa = 11.07GGEE400 pKa = 4.2GADD403 pKa = 3.33RR404 pKa = 11.84LYY406 pKa = 11.28GQGGDD411 pKa = 4.36DD412 pKa = 3.61VLIGQGGADD421 pKa = 3.75KK422 pKa = 11.16LVGADD427 pKa = 4.87GNDD430 pKa = 3.35TLYY433 pKa = 11.03GYY435 pKa = 10.91SDD437 pKa = 3.51NDD439 pKa = 3.55KK440 pKa = 10.99LYY442 pKa = 11.38GDD444 pKa = 4.5AGSDD448 pKa = 3.4KK449 pKa = 11.01LVGGDD454 pKa = 4.92GDD456 pKa = 3.77DD457 pKa = 3.57TLYY460 pKa = 11.36GGAGDD465 pKa = 4.13DD466 pKa = 4.17RR467 pKa = 11.84LYY469 pKa = 11.32ADD471 pKa = 4.43EE472 pKa = 5.4GADD475 pKa = 3.42NLYY478 pKa = 10.99GGTGADD484 pKa = 2.91TFVFTWLDD492 pKa = 3.56PSTLTSGGRR501 pKa = 11.84DD502 pKa = 3.57DD503 pKa = 5.18IYY505 pKa = 11.24QFSEE509 pKa = 4.02ADD511 pKa = 3.1GDD513 pKa = 4.2IINLSGIDD521 pKa = 3.91ANPTVNGGQAFDD533 pKa = 4.92FIGTNAFSEE542 pKa = 4.58QVSEE546 pKa = 3.92LRR548 pKa = 11.84YY549 pKa = 10.24VNTGSEE555 pKa = 4.26TYY557 pKa = 10.01IYY559 pKa = 11.06GDD561 pKa = 3.39INGDD565 pKa = 3.31GAADD569 pKa = 4.06FSIHH573 pKa = 6.01VDD575 pKa = 3.61GVVEE579 pKa = 4.06LQSIDD584 pKa = 4.05FILL587 pKa = 4.79
Molecular weight: 60.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W6W082|W6W082_9RHIZ Methyl-accepting chemotaxis sensory transducer OS=Rhizobium sp. CF080 OX=1144310 GN=PMI07_000986 PE=4 SV=1
MM1 pKa = 7.54TNRR4 pKa = 11.84LTRR7 pKa = 11.84RR8 pKa = 11.84LKK10 pKa = 10.56RR11 pKa = 11.84LAPQIIALAVILALITAVAPGFLNVSFQNGRR42 pKa = 11.84LYY44 pKa = 11.19GSLIDD49 pKa = 3.59ILVRR53 pKa = 11.84AAPVALLTIGMTLVIATRR71 pKa = 11.84GIDD74 pKa = 3.57LSIGAVIAICGAVAATLITHH94 pKa = 7.16GYY96 pKa = 7.69PLPVVILISLGVGLLCGLWNGVLVALLDD124 pKa = 3.7IQPIIATLILMVAGRR139 pKa = 11.84GIAQLITEE147 pKa = 4.47GVILTFNNDD156 pKa = 1.97SFAAVGSGSLAGIPIPILIWVCAALIIGLLVRR188 pKa = 11.84RR189 pKa = 11.84SALGFLIEE197 pKa = 3.95ATGINRR203 pKa = 11.84RR204 pKa = 11.84AATLAGVRR212 pKa = 11.84ARR214 pKa = 11.84FLLFFVYY221 pKa = 10.05AISGLCAAIAGLIVTADD238 pKa = 3.08IRR240 pKa = 11.84GADD243 pKa = 3.72ANNAGLWLEE252 pKa = 4.35LDD254 pKa = 4.62AILAVVIGGTSLNGGRR270 pKa = 11.84FSITASLIGALIIQSINTGILVSGFPPEE298 pKa = 3.76FNLIIKK304 pKa = 10.12AGIIMIVLTLQSPAIMALLGFVKK327 pKa = 10.5APRR330 pKa = 11.84RR331 pKa = 11.84KK332 pKa = 10.17AEE334 pKa = 3.96TTSRR338 pKa = 11.84EE339 pKa = 4.05TSHH342 pKa = 6.84KK343 pKa = 11.16AEE345 pKa = 4.03GTVRR349 pKa = 3.53
MM1 pKa = 7.54TNRR4 pKa = 11.84LTRR7 pKa = 11.84RR8 pKa = 11.84LKK10 pKa = 10.56RR11 pKa = 11.84LAPQIIALAVILALITAVAPGFLNVSFQNGRR42 pKa = 11.84LYY44 pKa = 11.19GSLIDD49 pKa = 3.59ILVRR53 pKa = 11.84AAPVALLTIGMTLVIATRR71 pKa = 11.84GIDD74 pKa = 3.57LSIGAVIAICGAVAATLITHH94 pKa = 7.16GYY96 pKa = 7.69PLPVVILISLGVGLLCGLWNGVLVALLDD124 pKa = 3.7IQPIIATLILMVAGRR139 pKa = 11.84GIAQLITEE147 pKa = 4.47GVILTFNNDD156 pKa = 1.97SFAAVGSGSLAGIPIPILIWVCAALIIGLLVRR188 pKa = 11.84RR189 pKa = 11.84SALGFLIEE197 pKa = 3.95ATGINRR203 pKa = 11.84RR204 pKa = 11.84AATLAGVRR212 pKa = 11.84ARR214 pKa = 11.84FLLFFVYY221 pKa = 10.05AISGLCAAIAGLIVTADD238 pKa = 3.08IRR240 pKa = 11.84GADD243 pKa = 3.72ANNAGLWLEE252 pKa = 4.35LDD254 pKa = 4.62AILAVVIGGTSLNGGRR270 pKa = 11.84FSITASLIGALIIQSINTGILVSGFPPEE298 pKa = 3.76FNLIIKK304 pKa = 10.12AGIIMIVLTLQSPAIMALLGFVKK327 pKa = 10.5APRR330 pKa = 11.84RR331 pKa = 11.84KK332 pKa = 10.17AEE334 pKa = 3.96TTSRR338 pKa = 11.84EE339 pKa = 4.05TSHH342 pKa = 6.84KK343 pKa = 11.16AEE345 pKa = 4.03GTVRR349 pKa = 3.53
Molecular weight: 36.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2077303 |
30 |
5677 |
311.4 |
33.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.616 ± 0.042 | 0.771 ± 0.009 |
5.608 ± 0.027 | 5.753 ± 0.026 |
3.965 ± 0.022 | 8.381 ± 0.027 |
1.977 ± 0.017 | 5.784 ± 0.027 |
3.791 ± 0.023 | 9.898 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.657 ± 0.015 | 2.892 ± 0.017 |
5.028 ± 0.022 | 3.068 ± 0.017 |
6.582 ± 0.03 | 5.825 ± 0.018 |
5.393 ± 0.026 | 7.379 ± 0.024 |
1.29 ± 0.012 | 2.343 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
