
Leucothrix arctica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Thiotrichaceae; Leucothrix
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
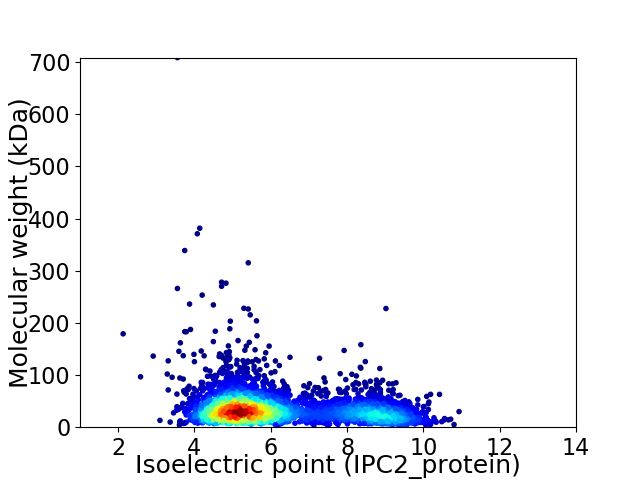
Virtual 2D-PAGE plot for 4192 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317CST0|A0A317CST0_9GAMM 3D-(3 5/4)-trihydroxycyclohexane-1 2-dione acylhydrolase (Decyclizing) OS=Leucothrix arctica OX=1481894 GN=iolD PE=3 SV=1
MM1 pKa = 7.83KK2 pKa = 10.24LRR4 pKa = 11.84PFLYY8 pKa = 10.19SATAMLSLNGAASAEE23 pKa = 4.55LIFSEE28 pKa = 4.6YY29 pKa = 10.28IEE31 pKa = 4.78GSGNNKK37 pKa = 9.7ALEE40 pKa = 4.52IYY42 pKa = 10.35NSSTADD48 pKa = 3.21VDD50 pKa = 3.87LSTYY54 pKa = 10.94SVVGYY59 pKa = 10.07QNGGADD65 pKa = 3.09AGYY68 pKa = 10.55SVDD71 pKa = 5.11LSGTLTAGSVYY82 pKa = 10.31IIAHH86 pKa = 6.05SSADD90 pKa = 3.55AEE92 pKa = 4.29ILAKK96 pKa = 10.24ADD98 pKa = 3.4QQYY101 pKa = 10.89GLQFNGDD108 pKa = 3.92DD109 pKa = 4.75AVTLQNSGTIVDD121 pKa = 4.52SIGKK125 pKa = 9.48IGEE128 pKa = 4.42DD129 pKa = 3.89PGSEE133 pKa = 3.79WGSGDD138 pKa = 3.75STTKK142 pKa = 10.73DD143 pKa = 2.84HH144 pKa = 6.16TLRR147 pKa = 11.84RR148 pKa = 11.84QAGLTTGDD156 pKa = 3.48TDD158 pKa = 3.54ATDD161 pKa = 4.75DD162 pKa = 4.81FDD164 pKa = 6.42PATQWDD170 pKa = 4.39AFEE173 pKa = 5.73KK174 pKa = 10.83DD175 pKa = 3.58DD176 pKa = 4.04FSGIGAHH183 pKa = 6.56GSLVVSDD190 pKa = 4.55GLPVANAGTDD200 pKa = 3.36QSGITAGAVVTLDD213 pKa = 3.33GSGSVDD219 pKa = 3.15TSEE222 pKa = 4.42GGSIIDD228 pKa = 4.02YY229 pKa = 10.68NWTQTSGTTVALDD242 pKa = 3.62TTDD245 pKa = 3.0PTMPSFTTPLAEE257 pKa = 5.53DD258 pKa = 3.93SLTFMLTVTDD268 pKa = 3.82NDD270 pKa = 4.09GNTATDD276 pKa = 3.57TVTVNVNAVLISQCNAQTHH295 pKa = 6.21VISQIQGSGDD305 pKa = 3.39TSPLVDD311 pKa = 3.63EE312 pKa = 4.87VVTVQAVVTKK322 pKa = 10.0LLPEE326 pKa = 3.87LTGYY330 pKa = 9.06MIQEE334 pKa = 4.0EE335 pKa = 4.35AADD338 pKa = 4.14EE339 pKa = 4.93DD340 pKa = 4.34GDD342 pKa = 4.03AATSEE347 pKa = 4.58GVFVYY352 pKa = 10.19DD353 pKa = 3.73YY354 pKa = 11.4ANSPSVGDD362 pKa = 3.54VVSIVGTVAEE372 pKa = 4.84YY373 pKa = 11.56YY374 pKa = 10.82DD375 pKa = 3.86LTQLKK380 pKa = 9.95SVSNFDD386 pKa = 3.45VCDD389 pKa = 3.49TGVSIAATDD398 pKa = 3.43ASLPFTAATDD408 pKa = 3.25IEE410 pKa = 4.96MYY412 pKa = 10.41EE413 pKa = 4.22GMLITFPQEE422 pKa = 3.57LTVTEE427 pKa = 4.77TYY429 pKa = 11.03SLGRR433 pKa = 11.84YY434 pKa = 7.7GQVALSSSGKK444 pKa = 9.93RR445 pKa = 11.84PNPTQVAAPGEE456 pKa = 4.22DD457 pKa = 3.24AVAQAAANALNLIIMDD473 pKa = 4.99DD474 pKa = 3.6GSNAQNATISYY485 pKa = 9.11PGFGEE490 pKa = 3.98LTYY493 pKa = 11.4DD494 pKa = 3.3NAIRR498 pKa = 11.84GGNTITGLTGVVNYY512 pKa = 7.84TFSTYY517 pKa = 11.0SVVPTADD524 pKa = 3.07VAFEE528 pKa = 4.8DD529 pKa = 4.59DD530 pKa = 3.76NARR533 pKa = 11.84TATPEE538 pKa = 3.97DD539 pKa = 3.29VGGEE543 pKa = 4.04LKK545 pKa = 11.1VSGFNVLNYY554 pKa = 7.5FTTLTSAGSEE564 pKa = 3.95ICGSGDD570 pKa = 3.42NYY572 pKa = 10.79CRR574 pKa = 11.84GADD577 pKa = 3.45DD578 pKa = 3.65AAEE581 pKa = 4.28FEE583 pKa = 4.33RR584 pKa = 11.84QNAKK588 pKa = 10.46LMSALSAIDD597 pKa = 3.78ADD599 pKa = 3.48IFGLIEE605 pKa = 3.97IEE607 pKa = 4.46NNAEE611 pKa = 3.84DD612 pKa = 4.09TAVSTLVEE620 pKa = 4.14NLNGEE625 pKa = 4.39VGADD629 pKa = 3.26TYY631 pKa = 11.27SYY633 pKa = 11.7VMTGMIGEE641 pKa = 4.58DD642 pKa = 3.76SIKK645 pKa = 10.29QALIYY650 pKa = 10.39KK651 pKa = 7.49PAKK654 pKa = 10.0VEE656 pKa = 3.9PFGDD660 pKa = 4.37FSIVDD665 pKa = 3.72YY666 pKa = 11.04NDD668 pKa = 3.2SKK670 pKa = 11.44NRR672 pKa = 11.84PSLIQTFKK680 pKa = 10.45TVGGEE685 pKa = 4.08DD686 pKa = 3.34EE687 pKa = 5.0DD688 pKa = 4.73AFTVVVNHH696 pKa = 6.68LKK698 pKa = 10.86SKK700 pKa = 10.87GSDD703 pKa = 3.41CDD705 pKa = 3.58EE706 pKa = 5.42LGDD709 pKa = 4.67PDD711 pKa = 6.07LEE713 pKa = 5.97DD714 pKa = 4.11GQGNCNLTRR723 pKa = 11.84LGAVEE728 pKa = 4.12TLLTSLNSMDD738 pKa = 3.79NQNILLLGDD747 pKa = 4.05FNSYY751 pKa = 11.07AMEE754 pKa = 4.68DD755 pKa = 4.33PITALKK761 pKa = 10.36DD762 pKa = 3.13AGYY765 pKa = 10.93VDD767 pKa = 5.14LGAEE771 pKa = 4.09AGEE774 pKa = 4.15TASYY778 pKa = 11.17VFDD781 pKa = 4.79GEE783 pKa = 4.34WGTLDD788 pKa = 3.49YY789 pKa = 11.51AFASASLADD798 pKa = 4.05KK799 pKa = 11.24VSDD802 pKa = 3.37TTEE805 pKa = 3.18WHH807 pKa = 6.53INADD811 pKa = 3.74EE812 pKa = 5.29PISFDD817 pKa = 3.58YY818 pKa = 9.86NTDD821 pKa = 3.77FKK823 pKa = 11.45ADD825 pKa = 3.72TEE827 pKa = 4.19IASYY831 pKa = 10.87YY832 pKa = 10.21GDD834 pKa = 3.47SAYY837 pKa = 10.4RR838 pKa = 11.84SSDD841 pKa = 3.32HH842 pKa = 7.34DD843 pKa = 3.84PVIVGLNITPVVDD856 pKa = 5.14DD857 pKa = 5.14GSNDD861 pKa = 4.84DD862 pKa = 6.09DD863 pKa = 7.04DD864 pKa = 7.25DD865 pKa = 7.33DD866 pKa = 6.58DD867 pKa = 5.6DD868 pKa = 6.0SSVGGGLAMSPLFMLFSLLGFAAIRR893 pKa = 11.84LRR895 pKa = 11.84RR896 pKa = 11.84KK897 pKa = 9.61
MM1 pKa = 7.83KK2 pKa = 10.24LRR4 pKa = 11.84PFLYY8 pKa = 10.19SATAMLSLNGAASAEE23 pKa = 4.55LIFSEE28 pKa = 4.6YY29 pKa = 10.28IEE31 pKa = 4.78GSGNNKK37 pKa = 9.7ALEE40 pKa = 4.52IYY42 pKa = 10.35NSSTADD48 pKa = 3.21VDD50 pKa = 3.87LSTYY54 pKa = 10.94SVVGYY59 pKa = 10.07QNGGADD65 pKa = 3.09AGYY68 pKa = 10.55SVDD71 pKa = 5.11LSGTLTAGSVYY82 pKa = 10.31IIAHH86 pKa = 6.05SSADD90 pKa = 3.55AEE92 pKa = 4.29ILAKK96 pKa = 10.24ADD98 pKa = 3.4QQYY101 pKa = 10.89GLQFNGDD108 pKa = 3.92DD109 pKa = 4.75AVTLQNSGTIVDD121 pKa = 4.52SIGKK125 pKa = 9.48IGEE128 pKa = 4.42DD129 pKa = 3.89PGSEE133 pKa = 3.79WGSGDD138 pKa = 3.75STTKK142 pKa = 10.73DD143 pKa = 2.84HH144 pKa = 6.16TLRR147 pKa = 11.84RR148 pKa = 11.84QAGLTTGDD156 pKa = 3.48TDD158 pKa = 3.54ATDD161 pKa = 4.75DD162 pKa = 4.81FDD164 pKa = 6.42PATQWDD170 pKa = 4.39AFEE173 pKa = 5.73KK174 pKa = 10.83DD175 pKa = 3.58DD176 pKa = 4.04FSGIGAHH183 pKa = 6.56GSLVVSDD190 pKa = 4.55GLPVANAGTDD200 pKa = 3.36QSGITAGAVVTLDD213 pKa = 3.33GSGSVDD219 pKa = 3.15TSEE222 pKa = 4.42GGSIIDD228 pKa = 4.02YY229 pKa = 10.68NWTQTSGTTVALDD242 pKa = 3.62TTDD245 pKa = 3.0PTMPSFTTPLAEE257 pKa = 5.53DD258 pKa = 3.93SLTFMLTVTDD268 pKa = 3.82NDD270 pKa = 4.09GNTATDD276 pKa = 3.57TVTVNVNAVLISQCNAQTHH295 pKa = 6.21VISQIQGSGDD305 pKa = 3.39TSPLVDD311 pKa = 3.63EE312 pKa = 4.87VVTVQAVVTKK322 pKa = 10.0LLPEE326 pKa = 3.87LTGYY330 pKa = 9.06MIQEE334 pKa = 4.0EE335 pKa = 4.35AADD338 pKa = 4.14EE339 pKa = 4.93DD340 pKa = 4.34GDD342 pKa = 4.03AATSEE347 pKa = 4.58GVFVYY352 pKa = 10.19DD353 pKa = 3.73YY354 pKa = 11.4ANSPSVGDD362 pKa = 3.54VVSIVGTVAEE372 pKa = 4.84YY373 pKa = 11.56YY374 pKa = 10.82DD375 pKa = 3.86LTQLKK380 pKa = 9.95SVSNFDD386 pKa = 3.45VCDD389 pKa = 3.49TGVSIAATDD398 pKa = 3.43ASLPFTAATDD408 pKa = 3.25IEE410 pKa = 4.96MYY412 pKa = 10.41EE413 pKa = 4.22GMLITFPQEE422 pKa = 3.57LTVTEE427 pKa = 4.77TYY429 pKa = 11.03SLGRR433 pKa = 11.84YY434 pKa = 7.7GQVALSSSGKK444 pKa = 9.93RR445 pKa = 11.84PNPTQVAAPGEE456 pKa = 4.22DD457 pKa = 3.24AVAQAAANALNLIIMDD473 pKa = 4.99DD474 pKa = 3.6GSNAQNATISYY485 pKa = 9.11PGFGEE490 pKa = 3.98LTYY493 pKa = 11.4DD494 pKa = 3.3NAIRR498 pKa = 11.84GGNTITGLTGVVNYY512 pKa = 7.84TFSTYY517 pKa = 11.0SVVPTADD524 pKa = 3.07VAFEE528 pKa = 4.8DD529 pKa = 4.59DD530 pKa = 3.76NARR533 pKa = 11.84TATPEE538 pKa = 3.97DD539 pKa = 3.29VGGEE543 pKa = 4.04LKK545 pKa = 11.1VSGFNVLNYY554 pKa = 7.5FTTLTSAGSEE564 pKa = 3.95ICGSGDD570 pKa = 3.42NYY572 pKa = 10.79CRR574 pKa = 11.84GADD577 pKa = 3.45DD578 pKa = 3.65AAEE581 pKa = 4.28FEE583 pKa = 4.33RR584 pKa = 11.84QNAKK588 pKa = 10.46LMSALSAIDD597 pKa = 3.78ADD599 pKa = 3.48IFGLIEE605 pKa = 3.97IEE607 pKa = 4.46NNAEE611 pKa = 3.84DD612 pKa = 4.09TAVSTLVEE620 pKa = 4.14NLNGEE625 pKa = 4.39VGADD629 pKa = 3.26TYY631 pKa = 11.27SYY633 pKa = 11.7VMTGMIGEE641 pKa = 4.58DD642 pKa = 3.76SIKK645 pKa = 10.29QALIYY650 pKa = 10.39KK651 pKa = 7.49PAKK654 pKa = 10.0VEE656 pKa = 3.9PFGDD660 pKa = 4.37FSIVDD665 pKa = 3.72YY666 pKa = 11.04NDD668 pKa = 3.2SKK670 pKa = 11.44NRR672 pKa = 11.84PSLIQTFKK680 pKa = 10.45TVGGEE685 pKa = 4.08DD686 pKa = 3.34EE687 pKa = 5.0DD688 pKa = 4.73AFTVVVNHH696 pKa = 6.68LKK698 pKa = 10.86SKK700 pKa = 10.87GSDD703 pKa = 3.41CDD705 pKa = 3.58EE706 pKa = 5.42LGDD709 pKa = 4.67PDD711 pKa = 6.07LEE713 pKa = 5.97DD714 pKa = 4.11GQGNCNLTRR723 pKa = 11.84LGAVEE728 pKa = 4.12TLLTSLNSMDD738 pKa = 3.79NQNILLLGDD747 pKa = 4.05FNSYY751 pKa = 11.07AMEE754 pKa = 4.68DD755 pKa = 4.33PITALKK761 pKa = 10.36DD762 pKa = 3.13AGYY765 pKa = 10.93VDD767 pKa = 5.14LGAEE771 pKa = 4.09AGEE774 pKa = 4.15TASYY778 pKa = 11.17VFDD781 pKa = 4.79GEE783 pKa = 4.34WGTLDD788 pKa = 3.49YY789 pKa = 11.51AFASASLADD798 pKa = 4.05KK799 pKa = 11.24VSDD802 pKa = 3.37TTEE805 pKa = 3.18WHH807 pKa = 6.53INADD811 pKa = 3.74EE812 pKa = 5.29PISFDD817 pKa = 3.58YY818 pKa = 9.86NTDD821 pKa = 3.77FKK823 pKa = 11.45ADD825 pKa = 3.72TEE827 pKa = 4.19IASYY831 pKa = 10.87YY832 pKa = 10.21GDD834 pKa = 3.47SAYY837 pKa = 10.4RR838 pKa = 11.84SSDD841 pKa = 3.32HH842 pKa = 7.34DD843 pKa = 3.84PVIVGLNITPVVDD856 pKa = 5.14DD857 pKa = 5.14GSNDD861 pKa = 4.84DD862 pKa = 6.09DD863 pKa = 7.04DD864 pKa = 7.25DD865 pKa = 7.33DD866 pKa = 6.58DD867 pKa = 5.6DD868 pKa = 6.0SSVGGGLAMSPLFMLFSLLGFAAIRR893 pKa = 11.84LRR895 pKa = 11.84RR896 pKa = 11.84KK897 pKa = 9.61
Molecular weight: 94.39 kDa
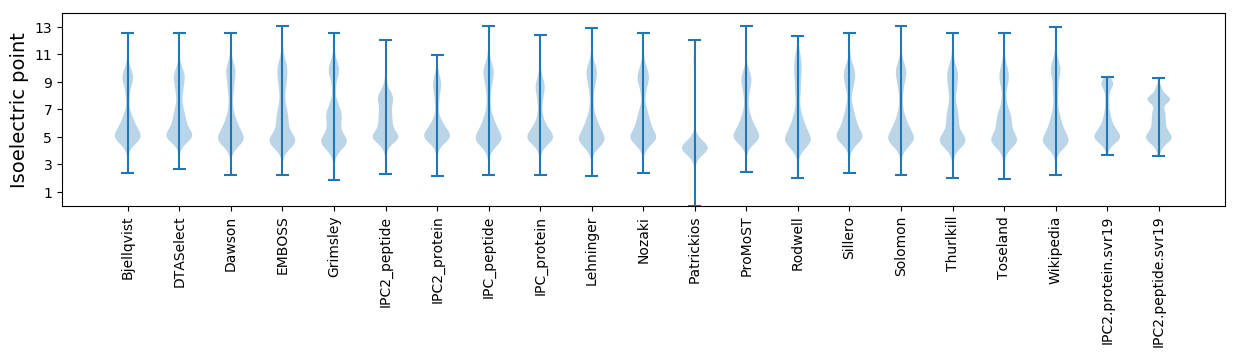
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317CFU3|A0A317CFU3_9GAMM Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase OS=Leucothrix arctica OX=1481894 GN=cobT PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.95GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTVGGRR28 pKa = 11.84KK29 pKa = 9.0VIKK32 pKa = 10.13ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84ALLCPP44 pKa = 4.17
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.95GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTVGGRR28 pKa = 11.84KK29 pKa = 9.0VIKK32 pKa = 10.13ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84ALLCPP44 pKa = 4.17
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1404415 |
18 |
6951 |
335.0 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.588 ± 0.047 | 0.96 ± 0.014 |
5.872 ± 0.043 | 6.115 ± 0.038 |
4.022 ± 0.031 | 6.937 ± 0.04 |
2.076 ± 0.022 | 6.343 ± 0.031 |
5.406 ± 0.041 | 10.195 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.518 ± 0.024 | 4.234 ± 0.029 |
3.964 ± 0.025 | 3.951 ± 0.032 |
4.486 ± 0.036 | 7.1 ± 0.042 |
5.88 ± 0.067 | 7.031 ± 0.035 |
1.262 ± 0.014 | 3.063 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
