
Common vole polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Microtus arvalis polyomavirus 1
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
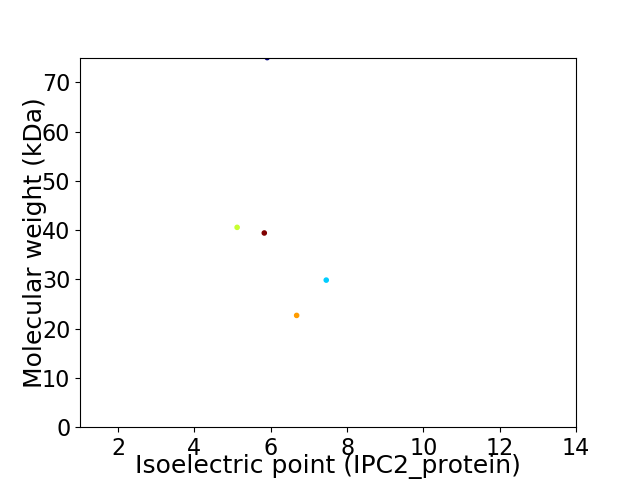
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0I7J3|A0A0P0I7J3_9POLY Major capsid protein VP1 OS=Common vole polyomavirus OX=1737523 PE=3 SV=1
MM1 pKa = 7.61GIVLALPEE9 pKa = 4.66FIAAAVAGGAEE20 pKa = 4.11ALEE23 pKa = 4.37IAGSVGALVEE33 pKa = 4.49GAADD37 pKa = 3.39TAAAIGTTAEE47 pKa = 4.22VLGEE51 pKa = 4.15SLPVTISEE59 pKa = 4.21AAATVLAQTPEE70 pKa = 4.05VVEE73 pKa = 4.06ALNAGGQAVGSAVSLGSGLYY93 pKa = 9.21QAISNTDD100 pKa = 3.61PQSGTHH106 pKa = 6.94GISDD110 pKa = 4.72RR111 pKa = 11.84IGMANALVPYY121 pKa = 9.38QQVPTYY127 pKa = 9.71LQWLRR132 pKa = 11.84NILSYY137 pKa = 10.76IPTVQQIWGGLEE149 pKa = 3.85RR150 pKa = 11.84ATQTYY155 pKa = 10.19QYY157 pKa = 10.95LDD159 pKa = 3.2EE160 pKa = 4.24QYY162 pKa = 11.19RR163 pKa = 11.84LRR165 pKa = 11.84EE166 pKa = 3.84QQYY169 pKa = 9.13EE170 pKa = 3.98RR171 pKa = 11.84LIAEE175 pKa = 4.61FEE177 pKa = 4.23RR178 pKa = 11.84GVNSGPVAGVIEE190 pKa = 5.2RR191 pKa = 11.84IRR193 pKa = 11.84ATALEE198 pKa = 4.4RR199 pKa = 11.84EE200 pKa = 4.57LEE202 pKa = 4.28VQRR205 pKa = 11.84IAGDD209 pKa = 3.51VRR211 pKa = 11.84DD212 pKa = 3.46VSTNLRR218 pKa = 11.84EE219 pKa = 4.19GLSASGRR226 pKa = 11.84AVANVGSALYY236 pKa = 8.68NTVSGAASATRR247 pKa = 11.84AAAGTLGGTVSNVVGHH263 pKa = 5.12VQGTISTIQDD273 pKa = 3.48YY274 pKa = 11.11LRR276 pKa = 11.84TIGSEE281 pKa = 3.71ASGVSTTMLTDD292 pKa = 4.77GYY294 pKa = 10.95NAMPRR299 pKa = 11.84GISNFGSWVFMSGSDD314 pKa = 3.28GGTPHH319 pKa = 6.94YY320 pKa = 11.02SLDD323 pKa = 3.45YY324 pKa = 8.27WVVYY328 pKa = 10.47ALEE331 pKa = 4.31EE332 pKa = 4.02ALKK335 pKa = 10.26PLYY338 pKa = 9.31STQNAPVEE346 pKa = 4.47STVEE350 pKa = 4.07DD351 pKa = 3.92EE352 pKa = 6.09DD353 pKa = 4.28DD354 pKa = 4.03TEE356 pKa = 4.35HH357 pKa = 7.11TPTKK361 pKa = 10.2NNKK364 pKa = 7.88KK365 pKa = 8.95RR366 pKa = 11.84RR367 pKa = 11.84RR368 pKa = 11.84RR369 pKa = 11.84SVVSRR374 pKa = 11.84PSKK377 pKa = 9.43RR378 pKa = 11.84RR379 pKa = 11.84HH380 pKa = 5.7RR381 pKa = 11.84SASS384 pKa = 3.25
MM1 pKa = 7.61GIVLALPEE9 pKa = 4.66FIAAAVAGGAEE20 pKa = 4.11ALEE23 pKa = 4.37IAGSVGALVEE33 pKa = 4.49GAADD37 pKa = 3.39TAAAIGTTAEE47 pKa = 4.22VLGEE51 pKa = 4.15SLPVTISEE59 pKa = 4.21AAATVLAQTPEE70 pKa = 4.05VVEE73 pKa = 4.06ALNAGGQAVGSAVSLGSGLYY93 pKa = 9.21QAISNTDD100 pKa = 3.61PQSGTHH106 pKa = 6.94GISDD110 pKa = 4.72RR111 pKa = 11.84IGMANALVPYY121 pKa = 9.38QQVPTYY127 pKa = 9.71LQWLRR132 pKa = 11.84NILSYY137 pKa = 10.76IPTVQQIWGGLEE149 pKa = 3.85RR150 pKa = 11.84ATQTYY155 pKa = 10.19QYY157 pKa = 10.95LDD159 pKa = 3.2EE160 pKa = 4.24QYY162 pKa = 11.19RR163 pKa = 11.84LRR165 pKa = 11.84EE166 pKa = 3.84QQYY169 pKa = 9.13EE170 pKa = 3.98RR171 pKa = 11.84LIAEE175 pKa = 4.61FEE177 pKa = 4.23RR178 pKa = 11.84GVNSGPVAGVIEE190 pKa = 5.2RR191 pKa = 11.84IRR193 pKa = 11.84ATALEE198 pKa = 4.4RR199 pKa = 11.84EE200 pKa = 4.57LEE202 pKa = 4.28VQRR205 pKa = 11.84IAGDD209 pKa = 3.51VRR211 pKa = 11.84DD212 pKa = 3.46VSTNLRR218 pKa = 11.84EE219 pKa = 4.19GLSASGRR226 pKa = 11.84AVANVGSALYY236 pKa = 8.68NTVSGAASATRR247 pKa = 11.84AAAGTLGGTVSNVVGHH263 pKa = 5.12VQGTISTIQDD273 pKa = 3.48YY274 pKa = 11.11LRR276 pKa = 11.84TIGSEE281 pKa = 3.71ASGVSTTMLTDD292 pKa = 4.77GYY294 pKa = 10.95NAMPRR299 pKa = 11.84GISNFGSWVFMSGSDD314 pKa = 3.28GGTPHH319 pKa = 6.94YY320 pKa = 11.02SLDD323 pKa = 3.45YY324 pKa = 8.27WVVYY328 pKa = 10.47ALEE331 pKa = 4.31EE332 pKa = 4.02ALKK335 pKa = 10.26PLYY338 pKa = 9.31STQNAPVEE346 pKa = 4.47STVEE350 pKa = 4.07DD351 pKa = 3.92EE352 pKa = 6.09DD353 pKa = 4.28DD354 pKa = 4.03TEE356 pKa = 4.35HH357 pKa = 7.11TPTKK361 pKa = 10.2NNKK364 pKa = 7.88KK365 pKa = 8.95RR366 pKa = 11.84RR367 pKa = 11.84RR368 pKa = 11.84RR369 pKa = 11.84SVVSRR374 pKa = 11.84PSKK377 pKa = 9.43RR378 pKa = 11.84RR379 pKa = 11.84HH380 pKa = 5.7RR381 pKa = 11.84SASS384 pKa = 3.25
Molecular weight: 40.58 kDa
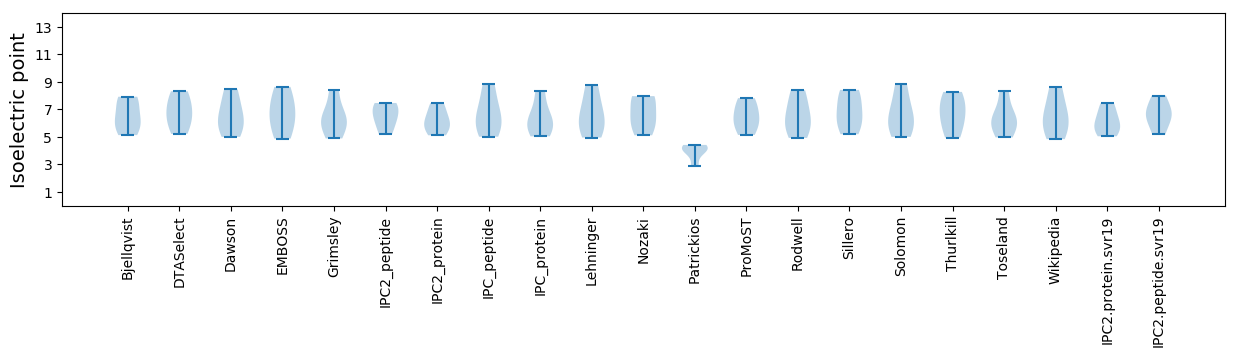
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0HGX9|A0A0P0HGX9_9POLY Large T antigen OS=Common vole polyomavirus OX=1737523 PE=4 SV=1
MM1 pKa = 7.56ANALVPYY8 pKa = 9.38QQVPTYY14 pKa = 9.71LQWLRR19 pKa = 11.84NILSYY24 pKa = 10.76IPTVQQIWGGLEE36 pKa = 3.85RR37 pKa = 11.84ATQTYY42 pKa = 10.19QYY44 pKa = 10.95LDD46 pKa = 3.2EE47 pKa = 4.24QYY49 pKa = 11.19RR50 pKa = 11.84LRR52 pKa = 11.84EE53 pKa = 3.84QQYY56 pKa = 9.13EE57 pKa = 3.98RR58 pKa = 11.84LIAEE62 pKa = 4.61FEE64 pKa = 4.23RR65 pKa = 11.84GVNSGPVAGVIEE77 pKa = 5.2RR78 pKa = 11.84IRR80 pKa = 11.84ATALEE85 pKa = 4.4RR86 pKa = 11.84EE87 pKa = 4.57LEE89 pKa = 4.28VQRR92 pKa = 11.84IAGDD96 pKa = 3.51VRR98 pKa = 11.84DD99 pKa = 3.46VSTNLRR105 pKa = 11.84EE106 pKa = 4.19GLSASGRR113 pKa = 11.84AVANVGSALYY123 pKa = 8.68NTVSGAASATRR134 pKa = 11.84AAAGTLGGTVSNVVGHH150 pKa = 5.12VQGTISTIQDD160 pKa = 3.48YY161 pKa = 11.11LRR163 pKa = 11.84TIGSEE168 pKa = 3.71ASGVSTTMLTDD179 pKa = 4.77GYY181 pKa = 10.95NAMPRR186 pKa = 11.84GISNFGSWVFMSGSDD201 pKa = 3.28GGTPHH206 pKa = 6.94YY207 pKa = 11.02SLDD210 pKa = 3.45YY211 pKa = 8.27WVVYY215 pKa = 10.47ALEE218 pKa = 4.31EE219 pKa = 4.02ALKK222 pKa = 10.26PLYY225 pKa = 9.31STQNAPVEE233 pKa = 4.47STVEE237 pKa = 4.07DD238 pKa = 3.92EE239 pKa = 6.09DD240 pKa = 4.28DD241 pKa = 4.03TEE243 pKa = 4.35HH244 pKa = 7.11TPTKK248 pKa = 10.2NNKK251 pKa = 7.88KK252 pKa = 8.95RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84SVVSRR261 pKa = 11.84PSKK264 pKa = 9.43RR265 pKa = 11.84RR266 pKa = 11.84HH267 pKa = 5.7RR268 pKa = 11.84SASS271 pKa = 3.25
MM1 pKa = 7.56ANALVPYY8 pKa = 9.38QQVPTYY14 pKa = 9.71LQWLRR19 pKa = 11.84NILSYY24 pKa = 10.76IPTVQQIWGGLEE36 pKa = 3.85RR37 pKa = 11.84ATQTYY42 pKa = 10.19QYY44 pKa = 10.95LDD46 pKa = 3.2EE47 pKa = 4.24QYY49 pKa = 11.19RR50 pKa = 11.84LRR52 pKa = 11.84EE53 pKa = 3.84QQYY56 pKa = 9.13EE57 pKa = 3.98RR58 pKa = 11.84LIAEE62 pKa = 4.61FEE64 pKa = 4.23RR65 pKa = 11.84GVNSGPVAGVIEE77 pKa = 5.2RR78 pKa = 11.84IRR80 pKa = 11.84ATALEE85 pKa = 4.4RR86 pKa = 11.84EE87 pKa = 4.57LEE89 pKa = 4.28VQRR92 pKa = 11.84IAGDD96 pKa = 3.51VRR98 pKa = 11.84DD99 pKa = 3.46VSTNLRR105 pKa = 11.84EE106 pKa = 4.19GLSASGRR113 pKa = 11.84AVANVGSALYY123 pKa = 8.68NTVSGAASATRR134 pKa = 11.84AAAGTLGGTVSNVVGHH150 pKa = 5.12VQGTISTIQDD160 pKa = 3.48YY161 pKa = 11.11LRR163 pKa = 11.84TIGSEE168 pKa = 3.71ASGVSTTMLTDD179 pKa = 4.77GYY181 pKa = 10.95NAMPRR186 pKa = 11.84GISNFGSWVFMSGSDD201 pKa = 3.28GGTPHH206 pKa = 6.94YY207 pKa = 11.02SLDD210 pKa = 3.45YY211 pKa = 8.27WVVYY215 pKa = 10.47ALEE218 pKa = 4.31EE219 pKa = 4.02ALKK222 pKa = 10.26PLYY225 pKa = 9.31STQNAPVEE233 pKa = 4.47STVEE237 pKa = 4.07DD238 pKa = 3.92EE239 pKa = 6.09DD240 pKa = 4.28DD241 pKa = 4.03TEE243 pKa = 4.35HH244 pKa = 7.11TPTKK248 pKa = 10.2NNKK251 pKa = 7.88KK252 pKa = 8.95RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84SVVSRR261 pKa = 11.84PSKK264 pKa = 9.43RR265 pKa = 11.84RR266 pKa = 11.84HH267 pKa = 5.7RR268 pKa = 11.84SASS271 pKa = 3.25
Molecular weight: 29.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1844 |
189 |
646 |
368.8 |
41.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.941 ± 1.522 | 2.223 ± 0.95 |
4.284 ± 0.527 | 7.646 ± 0.37 |
3.416 ± 0.852 | 6.941 ± 1.106 |
2.115 ± 0.558 | 4.826 ± 0.23 |
4.61 ± 1.355 | 8.785 ± 0.787 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.44 ± 0.411 | 5.043 ± 0.41 |
4.555 ± 0.84 | 4.393 ± 0.196 |
6.345 ± 0.589 | 6.996 ± 0.667 |
5.911 ± 0.982 | 7.05 ± 1.081 |
1.735 ± 0.379 | 3.742 ± 0.369 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
